3. Fundamentals II#
Course Website
%%html
<style> table {margin-left: 0 !important;} </style>
Scope#
flow equations
transport equations
generating flow nets (practical)
Readings#
Videos#
None
Spreadsheets/Programs/Data#
None
Flow Equations#
The governing equations for groundwater flow are derived by combining Darcy’s law—which describes how water moves through porous media under a hydraulic gradient—with the principle of mass conservation applied to a representative elementary volume (REV) of the aquifer. The result is a partial differential equation that relates changes in hydraulic head over time to the spatial variation in flow and any external sources or sinks of water. The specific form of the equation depends on the aquifer type, dimensionality, and storage properties.
Unconfined Aquifer:
For an unconfined aquifer, the saturated thickness of the aquifer changes as water levels rise and fall. In this case, transmissivity depends on the instantaneous head, so the conductivity terms are multiplied by \(h\). The left-hand side contains the product of the specific yield \(S_y\) and the rate of change of head \(\frac{\partial h}{\partial t}\), reflecting the volume of water released from storage per unit surface area per unit decline in head. The right-hand side sums the divergence of the flow in the \(x\) and \(y\) directions, and adds (or subtracts) source/sink terms for recharge, pumping, or leakage.
where:
\(S_y\) = specific yield.
Confined Aquifer (3D) :
In a confined aquifer, the saturated thickness remains constant, so transmissivity is independent of head. Instead, storage comes from the compressibility of the aquifer matrix and the pore water, represented in the specific storage term \(S_s =\rho g (\alpha + n \beta)\), where \(\alpha\) is the matrix compressibility, \(\beta\) is the fluid compressibility, \(n\) is porosity, \(\rho\) is fluid density, and \(g\) is gravitational acceleration. The fully three-dimensional form accounts for anisotropic conductivities \(K_x,K_y,K_z\) in all directions.
where:
\(S_s =\rho g (\alpha + n \beta)\).
\(\alpha\) = compressibility of porous matrix (a spring constant for the solids fraction).
\(\beta\) = compressibility of liquid (usually water).
Confined Aquifer (2D) :
When a confined aquifer can be treated in two dimensions—for example, in a horizontal flow model—the governing equation is vertically integrated over the aquifer thickness. The result is an equation in terms of transmissivity \(T\) and storage coefficient \(S\), where \(S\) is the vertical integral of \(S_s\) across the aquifer thickness. This 2D form is common in regional groundwater modeling where vertical gradients are small compared to horizontal gradients, and it simplifies computations while preserving the essential physics of lateral groundwater movement. A common extension is layers of 2D models with leakage between layers (quasi-3D).
where:
\(S\) = storage coefficient (\(S = \int_{z_{bot}}^{z_{top}}S_s(z) dz\)
Note
Origin of these equations
Starting Point : Darcy’s law + conservation of mass
Darcy’s law (vector form, possibly anisotropic):
\(𝑞=−𝐾 \nabla h\)
where
\(q\) is specific discharge
\(K\) is the hydraulic conductivity tensor
h is hydraulic head (a scalar field)
Continuity (single-phase water, small deformations):
\(\frac{\partial(\rho \theta)}{\partial t} + \nabla \cdot (\rho q) = \rho W\)
\(\theta\) is volumetric water content (\(\approx~\) porosity \(n\) when saturated), \(W\) is a volumetric source/sink per bulk volume (positive = recharge), and \(ρ\) is water density.
Assuming constant density (negligible variable-density effects) and rearranging:
\(\frac{\partial \theta}{\partial t} + \nabla \cdot q = W\)
All the physics now lives in (i) how \(\theta\) changes with \(h\) (storage) and (ii) \(q\) from Darcy’s law.
Confined aquifer (3D)
In a confined aquifer, the saturated thickness is constant; storage arises from compressibility of the matrix and water. A decline \(dh\) produces a volumetric water release \((S_s dh)\) per unit bulk volume, where
\(S_s =\rho g (\alpha + n \beta)\)
Here \(\alpha\) is matrix compressibility, \(\beta\) is water compressibility \(n\) is porosity, \(ρg\) converts head to pressure.
Thus
\(\frac{\partial \theta}{\partial t}~\approx~S_s\frac{\partial h}{\partial t}\)
Insert Darcy’s law \(𝑞=−𝐾 \nabla h\) as the equation of motion into continuity:
\(S_s\frac{\partial h}{\partial t}+ \nabla \cdot (−K \nabla h) = W \to S_s\frac{\partial h}{\partial t}= \nabla \cdot (K \nabla h) + W\)
which in components gives the 3D confined equation:
\(S_s \frac{\partial h}{\partial t} = \frac{\partial}{\partial x}(K_x\frac{\partial h}{\partial x}) + \frac{\partial}{\partial y}(K_y\frac{\partial h}{\partial y}) + \frac{\partial}{\partial z}(K_z\frac{\partial h}{\partial z})\pm W (source/sink)\)
Confined aquifer, vertically integrated (2D)
Integrate the 3D equation from \(z_{bot}\) to \(z_{top}\) (aquifer thickness \(B\). Define:
\(T_x = \int_{z_{bot}}^{z_{top}} K_x(z) dz~(\approx~K_xB)~~~~\) \(T_y = \int_{z_{bot}}^{z_{top}} K_y(z) dz~(\approx~K_yB)~~~~\) \(S = \int_{z_{bot}}^{z_{top}} S_s(z) dz~(\approx~S_sB)\)
Assuming \(h\) is nearly uniform with depth (Dupuit-type vertical averaging for confined systems), the vertical terms cancel on integration and sources/sinks convert to areal rates \(W'\). The result is:
\(S \frac{\partial h}{\partial t} = \frac{\partial}{\partial x}(T_x\frac{\partial h}{\partial x}) + \frac{\partial}{\partial y}(T_y\frac{\partial h}{\partial y}) \pm W' (source/sink)\)
which is the 2D confined equation.
Unconfined aquifer (Boussinesq form)
For unconfined conditions, the saturated thickness equals the head above the impermeable base (take datum \(z_b\)) so the thickness is \(b=h−z_b~\approx h ~\text{if}~z_b=0\). Two key changes:
Storage: draining/filling is dominated by specific yield \(S_y\) (drainable porosity), so
\(\frac{\partial \theta}{\partial t}~\approx~S_y \frac{\partial h}{\partial t}\).
Transmissivity depends on head: thickness \(b \simeq h\), so the horizontal fluxes use head-dependent transmissivity:
\(q_x=-K_x h \frac{\partial h}{\partial x}~~~~\),\(q_y=-K_y h \frac{\partial h}{\partial y}~~~~\),
after invoking a Dupuit assumption (vertical gradients small compared to horizontal; vertical head distribution is nearly hydrostatic).
Insert into continuity (plan-view):
\(S_y \frac{\partial h}{\partial t} = \frac{\partial}{\partial x}(K_x h \frac{\partial h}{\partial x}) + \frac{\partial}{\partial y}(K_y h \frac{\partial h}{\partial y}) \pm W' (source/sink)\)
which is the Boussinesq equation used for unconfined flow (the equation above, with optional sources/sinks).
Transport Equations#
Transport builds on the same foundations as flow: the equations of motion (Darcy’s law) and mass continuity. Here we add constituent-specific processes—advection and hydrodynamic dispersion—plus reactions (e.g., decay, biodegradation) and sorption. Although sorption is a reaction, it’s often treated separately because it directly alters storage via retardation.
We assume a saturated medium with constant fluid density and porosity over the time scales of interest. The velocity field is obtained from the groundwater flow solution described above (using \(\mathbf{q}\) or \(\mathbf{v}=\mathbf{q}/n\) as appropriate).
Variables and constitutive pieces#
\(C(\mathbf{x},t)\) — dissolved concentration \([M\,L^{-3}]\) in the mobile (water) phase
\(s(\mathbf{x},t)\) — sorbed concentration on the solid phase \([M\,M^{-1}]\)
\(n\) — porosity \([-]\); \(\ \rho_b\) — bulk density of porous medium \([M\,L^{-3}]\)
\(\mathbf{q}\) — Darcy flux (specific discharge) \([L\,T^{-1}]\)
\(\mathbf{v}=\mathbf{q}/n\) — average linear (pore) velocity \([L\,T^{-1}]\)
\(\boldsymbol{D}\) — hydrodynamic dispersion tensor \([L^{2}\,T^{-1}]\):\(~~~~ \boldsymbol{D} \;=\; \tau D_m\,\mathbf{I}\;+\; \alpha_T\,\lVert\mathbf{v}\rVert\,\mathbf{I}\;+\; (\alpha_L-\alpha_T)\,\frac{\mathbf{v}\mathbf{v}^{\!\top}}{\lVert\mathbf{v}\rVert}\) where \(D_m\) is molecular diffusion, \(\tau\) is tortuosity, and \(\alpha_L,\alpha_T\) are longitudinal/transverse dispersivities.
\(W(\mathbf{x},t)\) — volumetric water source/sink per bulk volume \([T^{-1}]\) (positive = water addition)
\(C_{\mathrm{in}}(\mathbf{x},t)\) — concentration of water added through \(W>0\)
Dissolved mass flux (dispersion + advection): $\( \mathbf{J} \;=\; -\,n\,\boldsymbol{D}\,\nabla C \;+\; \mathbf{q}\,C \qquad [M\,L^{-2}\,T^{-1}]. \)$
Species mass balance over a representative elementary volume (REV)#
Total species mass per bulk volume: $\( M \;=\; nC \;+\; \rho_b s. \)$
Conservation of mass (accumulation = inflow \(-\) outflow \(+\) reactions \(+\) external additions): $\( \frac{\partial}{\partial t}\big(nC + \rho_b s\big) \;+\; \nabla\!\cdot\mathbf{J} \;=\; nW\,C_{\mathrm{in}} \;+\; R_{\text{chem}}(C,s,\ldots), \)\( where \)R_{\text{chem}}\( \)[M,L^{-3},T^{-1}]$ is net internal production/consumption from reactions not otherwise represented (e.g., multispecies kinetics, biodegradation).
Insert \(\mathbf{J}\): $\( \frac{\partial}{\partial t}\big(nC + \rho_b s\big) \;+\; \nabla\!\cdot\!\big(-\,n\boldsymbol{D}\nabla C + \mathbf{q}C\big) \;=\; nW\,C_{\mathrm{in}} \;+\; R_{\text{chem}}. \)$
Using groundwater continuity, \(\nabla\!\cdot\mathbf{q}=nW\), we have
\(-\nabla\!\cdot(\mathbf{q}C)= -\mathbf{q}\!\cdot\nabla C \;-\; C\,\nabla\!\cdot\mathbf{q} = -\mathbf{q}\!\cdot\nabla C - nW\,C\).
Together with \(+\,nW\,C_{\mathrm{in}}\), the net water exchange term is \(nW(C_{\mathrm{in}}-C)\).
Sorption and retardation (a type of reaction)#
(a) Instantaneous linear equilibrium sorption#
Assume \(s = K_d\,C\). Then $\( \frac{\partial}{\partial t}(nC + \rho_b s) = \big(n + \rho_b K_d\big)\,\frac{\partial C}{\partial t} = n\,R_f\,\frac{\partial C}{\partial t}, \qquad R_f \equiv 1 + \frac{\rho_b K_d}{n}. \)$
Substitute into the balance equation above:
where \(-\lambda n C\) represents first-order aqueous decay (optional).
Divide by \(n\) and use \(\mathbf{v}=\mathbf{q}/n\) if preferred:
(b) Kinetic sorption (first-order two-rate; optional)#
Replace \(s=K_d C\) with $\( \rho_b\frac{\partial s}{\partial t} = k_a\,\rho_b C \;-\; k_d\,\rho_b s, \)\( and keep \)\rho_b ,\partial s/\partial t\( explicitly in the accumulation term. No single \)R_f$ applies; this yields a coupled PDE–ODE system.
(c) Dual-porosity / mobile–immobile (optional)#
Let \(n = n_m + n_{im}\) and introduce \(C_{im}\): $\( \begin{aligned} & n_m\frac{\partial C}{\partial t} + \rho_b\frac{\partial s}{\partial t} \;-\; \nabla\!\cdot\!\big(n_m\boldsymbol{D}\nabla C\big) + \nabla\!\cdot(\mathbf{q}C) = nW\,C_{\mathrm{in}} + R_{\text{chem}} - \omega\,(C_{im}-C) - \lambda\,n_m C, \\[4pt] & n_{im}\frac{\partial C_{im}}{\partial t} = \omega\,(C_{im}-C) - \lambda_{im}\,n_{im} C_{im}. \end{aligned} \)$
Canonical forms (for direct use)#
General conservative form (no assumption on sorption kinetics): $\( \frac{\partial}{\partial t}\big(nC + \rho_b s\big) = \nabla\!\cdot\!\big(n\boldsymbol{D}\nabla C\big) - \nabla\!\cdot(\mathbf{q}C) + nW\,C_{\mathrm{in}} + R_{\text{chem}}(C,s,\ldots) - \lambda\,nC. \)$
Retarded ADE with first-order decay (instantaneous linear sorption): $\( nR_f\,\frac{\partial C}{\partial t} = \nabla\!\cdot\!\big(n\boldsymbol{D}\nabla C\big) - \nabla\!\cdot(\mathbf{q}C) + nW\,C_{\mathrm{in}} + R_{\text{chem}}(C) - \lambda\,nC. \)$
(Optionally divide by \(n\) and write in terms of \(\mathbf{v}=\mathbf{q}/n\).)
Boundary conditions (quick reference)#
Dirichlet (specified concentration): \(C = C_b\) on \(\Gamma_D\).
Neumann (specified mass flux): \(\mathbf{n}\!\cdot\!\big(-n\boldsymbol{D}\nabla C + \mathbf{q}C\big) = J_b\) on \(\Gamma_N\).
No-flux: \(\mathbf{n}\!\cdot\!\big(-n\boldsymbol{D}\nabla C + \mathbf{q}C\big)=0\).
Units and sign checks#
\(\nabla\!\cdot(n\boldsymbol{D}\nabla C)\) and \(-\nabla\!\cdot(\mathbf{q}C)\) both have units \([M\,L^{-3}\,T^{-1}]\).
\(nW(C_{\mathrm{in}}-C)\) has units \([M\,L^{-3}\,T^{-1}]\).
Positive \(W\) adds water and mass; negative \(W\) removes them.
Notes.
(i) If density varies or saturation changes, retain \(\rho\) and use the appropriate flow equation (e.g., Richards’ equation) and variable-density transport.
(ii) Spatially variable \(n(\mathbf{x})\) can be kept inside divergences; if you divide by \(n\), handle \(\nabla n\) terms consistently.
Solving the Equations#
Flow and transport are typically solved on the same spatial domain but with different numerical behaviors. Groundwater flow is elliptic at steady state and parabolic when storage is included; mature solvers (finite difference/volume/element with implicit time stepping) handle these problems reliably. By contrast, transport (advection–dispersion–reaction) mixes hyperbolic advection with parabolic dispersion and adds reactions, which makes it more prone to numerical artifacts (numerical dispersion, spurious oscillations, and CFL-related stability limits). As a result, transport often needs stabilization (e.g., upwinding/TVD/SUPG), careful grid design, and step-size control.
Where geometry and boundary conditions are simple, analytical solutions exist and are preferred for verification and insight—we will use several later. For realistic sites, we use numerical models: in this course we’ll rely on MODFLOW 6 for flow (with FloPy to script workflows), remembering that the governing equations are precisely those stated above. When reactions matter (sorption, decay, multispecies kinetics), a common and practical strategy is operator splitting: advance transport for a short step, then apply reaction kinetics locally (cell-by-cell), update concentrations, and iterate. Fully coupled (monolithic) reactive-transport solvers also exist but are more specialized. In practice, many workflows compose reliable codes via APIs—a flow solver, a transport stepper, and a reaction module—to simulate multiple interacting species without re-implementing everything from scratch.
To build intuition before the full machinery, the next section generates numerical flownets. These approximate head fields and streamlines under idealized conditions, making visible how boundary conditions, heterogeneity, and anisotropy shape flow patterns. The flownets give a concrete feel for what the flow solver is doing and set the stage for interpreting transport results that follow.
Flownets#
A flow net is a graphical representation that depicts the flow of groundwater within an aquifer or through porous media. It consists of a network of flow lines and equipotential lines that together provide a clear visualization of the groundwater flow direction and hydraulic head distribution within a given geological formation. Flow lines represent the path that groundwater particles would take as they move through the aquifer, while equipotential lines represent lines of equal hydraulic head. These lines intersect orthogonally, forming a grid-like pattern, with the density of flow lines increasing where the groundwater flow is faster.
Flow nets are employed in various aspects of groundwater modeling:
Delineating Flow Patterns: Flow nets are instrumental in identifying the direction and velocity of groundwater flow within an aquifer. By studying the flow lines and equipotential lines, hydrogeologists can determine the dominant flow pathways and areas of groundwater recharge and discharge.
Designing Well Fields: When designing well fields for groundwater extraction, it is crucial to understand the local groundwater flow patterns. Flow nets help in optimizing the placement and pumping rates of wells, ensuring efficient water supply without causing adverse effects on nearby groundwater-dependent ecosystems.
Contaminant Transport Modeling: Flow nets are an essential component of contaminant transport modeling. They provide the basis for predicting the movement of contaminants through the subsurface, allowing for the assessment of potential risks and the development of remediation strategies.
Dam and Barrier Design: Engineers use flow nets to design dams, levees, and groundwater barriers. Understanding groundwater flow patterns is critical for ensuring the stability and effectiveness of these structures in controlling water flow.
Land Use Planning: Flow nets also have applications in land use planning. By considering groundwater flow patterns, urban planners can make informed decisions about where to build infrastructure, manage stormwater, and protect sensitive areas from potential contamination.
Flownets provide a visual representation of groundwater flow patterns and hydraulic head distributions, aiding in various aspects of groundwater resource management, environmental protection, and engineering design.
Numerical generation of flownets can be accomplished using a spreadsheet, python script, or MODFLOW.
A simple spreadsheet example is available from
Cleveland, T.G. (1997) Velocity Potential - Stream Function Ideal Flow Model (circa 1999) (Excel)
Example in Python#
In steady aquifer flow, the flow is irrotational (or at least can be modeled as such). There exists an orthogonal function called the stream function (it is the function that exists in the flow field when vorticity is zero). A really good explaination of stream functions (and streamlines) appears on pages 381–398 in Zheng(1995).
This function can be used to generate plots of streamlines for the same system. The principal changes are the material properties representation and the boundary conditions.
Velocity Potential#
The velocity potential function is usually Darcy’s law applied to the head distribution \(\phi = \frac{T}{b}h\)
If we start with a steady flow description of GW flow like:
the terms \({\frac{T_x}{b} \frac{\partial h}{\partial x}}\) and \({\frac{T_y}{b} \frac{\partial h}{\partial y}}\) can be replaced by \(\frac{\partial \phi}{\partial x}\) or \(\frac{\partial \phi}{\partial y}\) so the expression looks something like
Which is what we would call the velocity potential - although for flow nets we usually just solve for the head so we can compute pressures, using the formulation below:
The difference equation (pp. 349-357) on a regular grid is
Typically one approximates the spatial variation of the material properties (transmissivity) as arithmetic mean values between two cells, so making the following definitions:
Substitution into the difference equation yields
As before we can explicitly write the cell equation for \(h_{i,j}\) as
This difference equation represents an approximation to the governing flow equation, the accuracy depending on the cell size. Boundary conditions are applied directly into the analogs (another name for the difference equations) at appropriate locations on the computational grid. We can generate solutions either by iteration or solving the resulting linear system; iteration is really easy to program, so for exploratory work that’s usually the approach.
Stream Function#
The stream function \(\Psi\) is a function that is orthogonal to the velocity potential function and expressed as a partial differential equation is
Observe how the material property is inverted and changes directional identity, otherwise the equation is structurally identical to the groundwater flow equation (for steady flow).
The difference equation is practically the same as above:
The difference equation is also almost the same
The substitutions are
Substitution into the difference equation yields
As before we can explicitly write the cell equation for \(h_{i,j}\) as
So at this point we could literally use the same script, however boundary conditions also ``invert.’’ A no-flow head-domain boundary is a constant value stream function-domain boundary. A constant value head-domain boundary is a zero-gradient stream function-domain boundary.
So a script that first solves for the head distribution, then the stream function could be used to create a flownet.
Sheetpile under a dam flownet#
Compare to the example shown on pp. 30-34.
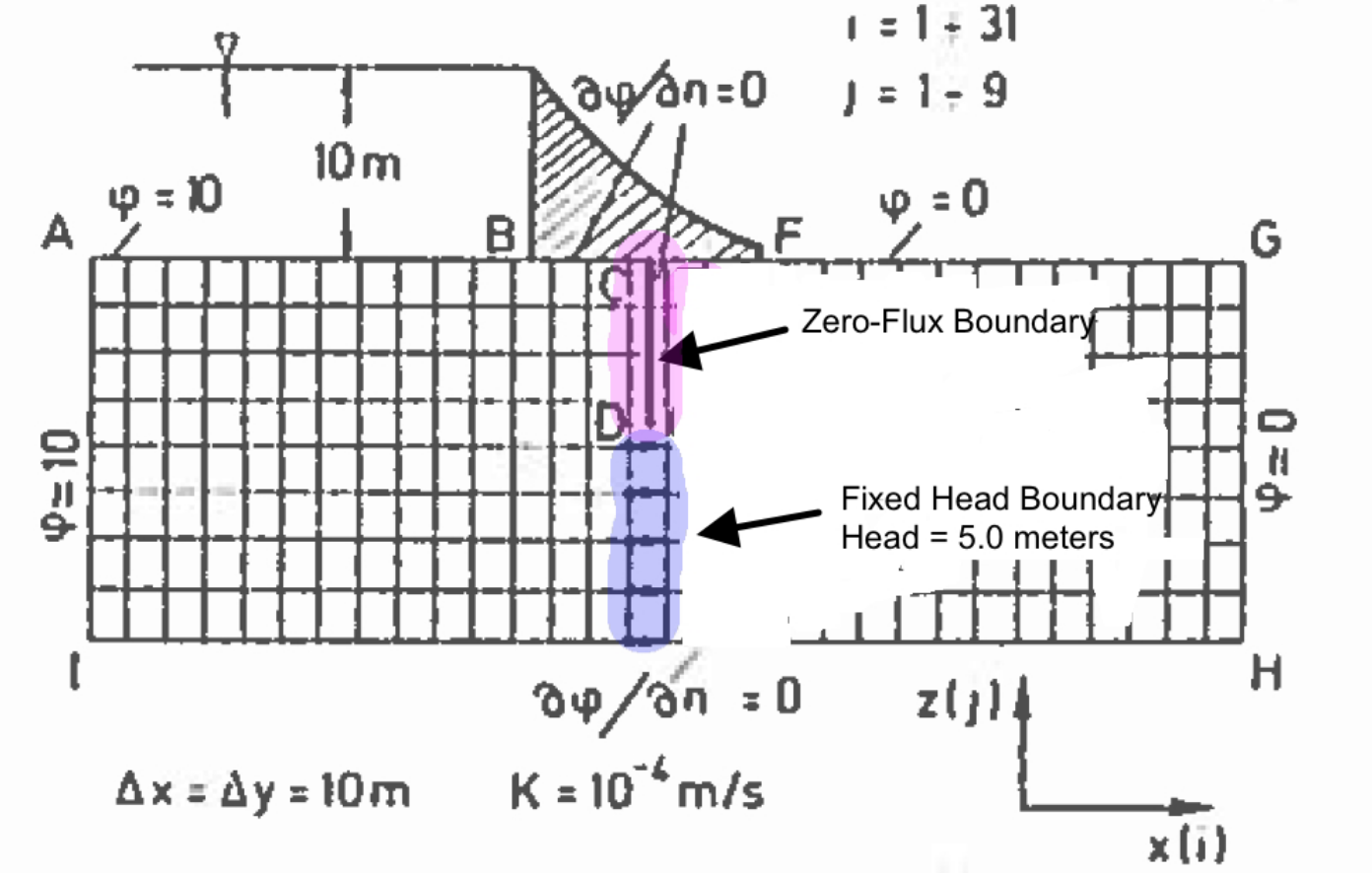
The figure below is a schematic of a dam built upon a permeable ground layer 80 meters thick (segment A to I).
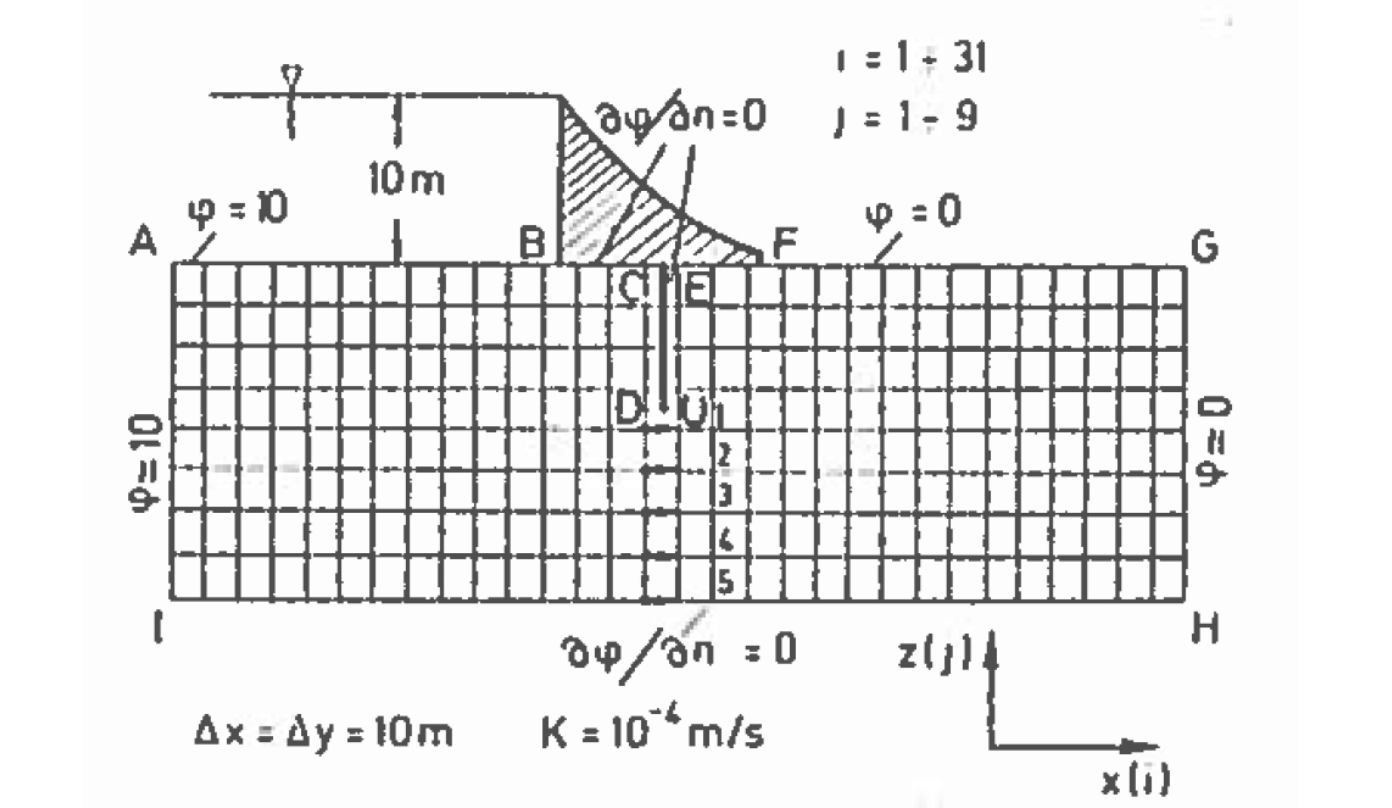
The dam has a base 60 meters long (segment B to F), with an upstream water depth of 10 meters. The downstream side of the dam is at 0 meters depth (otherwise its not a very good dam!). A sheetpile cutoff wall is installed beneath the dam (segment C to D to U to E). The ground layer has a hydraulic conductivity of K = 1 × 10−4 meters per second.
Important engineering questions are what is the pore water pressure under the dam, and is what is the total seepage under the dam? The pore water pressure can be found by solving for heads under the dam then subtracting the elevation of the computation location relative to a datum. The flow is found by Darcy’s law applied under the dam (shown as locations 1,2,3,4, and 5 in the sketch), which in turn requires computation of head under the dam. Thus the questions are answered by finding the head distribution under the dam.
The flow field (mathematically) extends an infinite distance upstream and downstream, but as a practical matter the contribution to seepage far upstream of the dam is negligible, and hence is approximated by the finite domain depicted.
Using the tools we have already we can simply build an input file, run our script and determine the head distribution (and thus compute the discharges under the dam. Then rerun with same script, but modify the boundary conditions and material properties to obtain the stream function - plot both solutions on the same contour map and viola we have a flownet.
The first is to represent the domain as shown, and make the following specifications in the boundary condition information, and we will treat the sheetpile cutoff wall as a low permeability inclusion (much like the the prior example). The boundary conditions are:
The segment from A to B is a constant head boundary with value equal to 10.
The segment from B to F is a zero-flux boundary.
The segment from B to C to D to U to E to F should be treated as a zero-flux boundary, but our mask does not extend into the interior – however the sheetpile itself can be approximated by providing a very small permeability. Alternately we could (should) modify the code to handle interior boundaries – but that is outside the scope of this chapter.
The segment from F to G is a constant head boundary with value equal to 0.
The segment form G to H is a constant head boundary with value equal to 0.
The segment from H to I is a zero-flux boundary.
The segment from I to A is a constant head boundary with value equal to 10.
Here’s the input file for the heads \(\phi\) in the picture
10
10
1
9
31
1e-12
5000
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 180 190 200 210 220 230 240 250 260 270 280 290 300
80 70 60 50 40 30 20 10 0
1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1 1 1 1 1 1 1 1 1
1 1 1 1 1 1 1 1 1
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0000001 0.0000001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0000001 0.0000001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0000001 0.0000001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0000001 0.0000001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0000001 0.0000001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.00010
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0000001 0.0000001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0000001 0.0000001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0000001 0.0000001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0000001 0.0000001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0000001 0.0000001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
def sse(matrix1,matrix2):
sse=0.0
nr=len(matrix1) # get row count
nc=len(matrix1[0]) # get column count
for ir in range(nr):
for jc in range(nc):
sse=sse+(matrix1[ir][jc]-matrix2[ir][jc])**2
return(sse)
def update(matrix1,matrix2):
nr=len(matrix1) # get row count
nc=len(matrix1[0]) # get column count
##print(nr,nc)
for ir in range(nr):
for jc in range(nc):
##print(ir,jc)
matrix2[ir][jc]=matrix1[ir][jc]
return(matrix2)
def writearray(matrix):
nr=len(matrix) # get row count
nc=len(matrix[0]) # get column count
import numpy as np
new_list = list(np.around(np.array(matrix), 3))
for ir in range(nr):
print(ir,new_list[ir][:])
return()
localfile = open("PotentialFnSheetpile1.txt","r") # connect and read file for 2D gw model
deltax = float(localfile.readline())
deltay = float(localfile.readline())
deltaz = float(localfile.readline())
nrows = int(localfile.readline())
ncols = int(localfile.readline())
tolerance = float(localfile.readline())
maxiter = int(localfile.readline())
distancex = [] # empty list
distancex.append([float(n) for n in localfile.readline().strip().split()])
distancey = [] # empty list
distancey.append([float(n) for n in localfile.readline().strip().split()])
boundarytop = [] #empty list
boundarytop.append([float(n) for n in localfile.readline().strip().split()])
boundarybottom = [] #empty list
boundarybottom.append([int(n) for n in localfile.readline().strip().split()])
boundaryleft = [] #empty list
boundaryleft.append([int(n) for n in localfile.readline().strip().split()])
boundaryright = [] #empty list
boundaryright.append([int(n) for n in localfile.readline().strip().split()])
head =[] # empty list
for irow in range(nrows):
head.append([float(n) for n in localfile.readline().strip().split()])
hydcondx = [] # empty list
for irow in range(nrows):
hydcondx.append([float(n) for n in localfile.readline().strip().split()])
hydcondy = [] # empty list
for irow in range(nrows):
hydcondy.append([float(n) for n in localfile.readline().strip().split()])
localfile.close() # Disconnect the file
##
amat = [[0 for j in range(ncols)] for i in range(nrows)]
bmat = [[0 for j in range(ncols)] for i in range(nrows)]
cmat = [[0 for j in range(ncols)] for i in range(nrows)]
dmat = [[0 for j in range(ncols)] for i in range(nrows)]
##
for irow in range(1,nrows-1):
for jcol in range(1,ncols-1):
amat[irow][jcol] = (( hydcondx[irow-1][jcol ]+ hydcondx[irow ][jcol ]) * deltaz ) /(2.0*deltax**2)
bmat[irow][jcol] = (( hydcondx[irow ][jcol ]+ hydcondx[irow+1][jcol ]) * deltaz ) /(2.0*deltax**2)
cmat[irow][jcol] = (( hydcondy[irow ][jcol-1]+ hydcondy[irow ][jcol ]) * deltaz ) /(2.0*deltay**2)
dmat[irow][jcol] = (( hydcondy[irow ][jcol ]+ hydcondy[irow ][jcol+1]) * deltaz ) /(2.0*deltay**2)
##
headold = [[0 for jc in range(ncols)] for ir in range(nrows)] #force new matrix
headold = update(head,headold) # update
##writearray(head)
##print("----")
##writearray(headold)
##print("--------")
tolflag = False
for iter in range(maxiter):
## print("begin iteration")
## writearray(head)
## print("----")
## writearray(headold)
## print("--------")
# Boundary Conditions
# first and last row special == no flow boundaries
for jcol in range(ncols):
if boundarytop[0][jcol] == 0: # no - flow at top
head [0][jcol ] = head[1][jcol ]
if boundarybottom[0][ jcol ] == 0: # no - flow at bottom
head [nrows-1][jcol ] = head[nrows-2][jcol ]
# first and last column special == no flow boundaries
for irow in range(nrows):
if boundaryleft[0][ irow ] == 0:
head[irow][0] = head [irow][1] # no - flow at left
if boundaryright[0][ irow ] == 0:
head [irow][ncols-1] = head[ irow ][ncols-2] # no - flow at right
# interior updates
for irow in range(1,nrows-1):
for jcol in range(1,ncols-1):
head[irow][jcol]=( amat[irow][jcol]*head[irow-1][jcol ]
+bmat[irow][jcol]*head[irow+1][jcol ]
+cmat[irow][jcol]*head[irow ][jcol-1]
+dmat[irow][jcol]*head[irow ][jcol+1])/(amat[ irow][jcol ]+ bmat[ irow][jcol ]+ cmat[ irow][jcol ]+ dmat[ irow][jcol ])
# test for stopping iterations
## print("end iteration")
## writearray(head)
## print("----")
## writearray(headold)
percentdiff = sse(head,headold)
if percentdiff <= tolerance:
print("Exit iterations in velocity potential because tolerance met ")
print("Iterations =" , iter+1 ) ;
tolflag = True
break
headold = update(head,headold)
print("End Calculations")
print("Iterations = ",iter+1)
print("Closure Error = ",round(percentdiff,3))
print("Head Map")
print("----")
writearray(head)
print("----")
############# Contour Plot
my_xyz = [] # empty list
for irow in range(nrows):
for jcol in range(ncols):
my_xyz.append([distancex[0][jcol],distancey[0][irow],head[irow][jcol]])
import pandas
my_xyz = pandas.DataFrame(my_xyz) # convert into a data frame
#print(my_xyz) # activate to examine the dataframe
import numpy
import matplotlib.pyplot
from scipy.interpolate import griddata
# extract lists from the dataframe
coord_x = my_xyz[0].values.tolist() # column 0 of dataframe
coord_y = my_xyz[1].values.tolist() # column 1 of dataframe
coord_z = my_xyz[2].values.tolist() # column 2 of dataframe
coord_xy = numpy.column_stack((coord_x, coord_y))
# Set plotting range in original data units
lon = numpy.linspace(min(coord_x), max(coord_x), 3000)
lat = numpy.linspace(min(coord_y), max(coord_y), 800)
X, Y = numpy.meshgrid(lon, lat)
# Grid the data; use linear interpolation (choices are nearest, linear, cubic)
Z = griddata(numpy.array(coord_xy), numpy.array(coord_z), (X, Y), method='cubic')
# Build the map
fig, ax = matplotlib.pyplot.subplots()
fig.set_size_inches(15, 4)
levels1 = [1,2,3,4,5,6,7,8,9,10]
CS = ax.contour(X, Y, Z, levels1)
ax.clabel(CS, inline=2, fontsize=16)
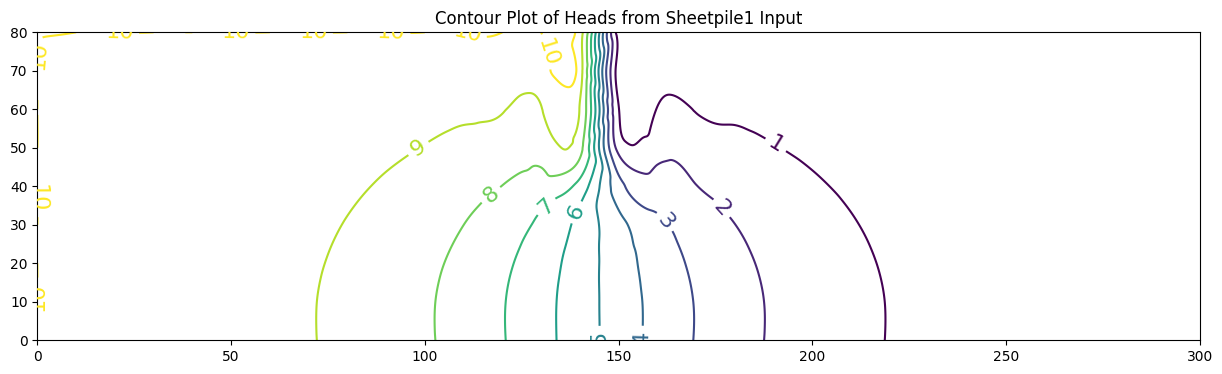
ax.set_title('Contour Plot of Heads from Sheetpile1 Input')
Exit iterations in velocity potential because tolerance met
Iterations = 443
End Calculations
Iterations = 443
Closure Error = 0.0
Head Map
----
0 [10. 10. 10. 10. 10. 10. 10. 10. 10. 10.
10. 10. 10. 10. 9.478 0.524 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. ]
1 [10. 9.98 9.959 9.936 9.91 9.881 9.848 9.808 9.763 9.71
9.652 9.591 9.535 9.497 9.478 0.524 0.505 0.467 0.411 0.351
0.293 0.241 0.196 0.157 0.125 0.097 0.073 0.052 0.034 0.016
0. ]
2 [10. 9.96 9.919 9.874 9.824 9.767 9.701 9.623 9.532 9.426
9.306 9.178 9.054 8.965 8.949 1.055 1.039 0.95 0.827 0.699
0.58 0.475 0.385 0.309 0.244 0.19 0.143 0.102 0.066 0.032
0. ]
3 [10. 9.943 9.883 9.818 9.746 9.663 9.566 9.451 9.316 9.156
8.969 8.759 8.538 8.351 8.336 1.67 1.654 1.468 1.247 1.038
0.852 0.694 0.56 0.448 0.354 0.275 0.207 0.148 0.095 0.047
0. ]
4 [10. 9.927 9.852 9.77 9.678 9.572 9.448 9.3 9.124 8.912
8.657 8.351 7.986 7.559 6.629 3.379 2.448 2.021 1.657 1.352
1.098 0.888 0.714 0.57 0.45 0.348 0.262 0.187 0.121 0.059
0. ]
5 [10. 9.915 9.827 9.731 9.623 9.499 9.353 9.179 8.968 8.711
8.395 8.002 7.498 6.806 5.702 4.307 3.202 2.511 2.007 1.615
1.301 1.046 0.838 0.667 0.526 0.407 0.306 0.219 0.141 0.069
0. ]
6 [10. 9.907 9.81 9.704 9.586 9.448 9.287 9.093 8.858 8.569
8.212 7.764 7.196 6.465 5.528 4.48 3.544 2.813 2.246 1.8
1.444 1.158 0.926 0.736 0.579 0.448 0.337 0.24 0.155 0.076
0. ]
7 [10. 9.903 9.801 9.69 9.566 9.422 9.253 9.049 8.801 8.496
8.118 7.647 7.057 6.33 5.467 4.542 3.679 2.952 2.364 1.894
1.518 1.215 0.971 0.771 0.606 0.469 0.352 0.251 0.162 0.079
0. ]
8 [10. 9.903 9.801 9.69 9.566 9.422 9.253 9.049 8.801 8.496
8.118 7.647 7.057 6.33 5.467 4.542 3.679 2.952 2.364 1.894
1.518 1.215 0.971 0.771 0.606 0.469 0.352 0.251 0.162 0.079
0. ]
----
Text(0.5, 1.0, 'Contour Plot of Heads from Sheetpile1 Input')
Now we reuse the code, but change head to stream and modify the contour plot variable names as well as modify the input file to reflect the changed material properties and boundary conditions.
Here’s the input file for the stream function
10
10
1
9
31
1e-12
5000
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 180 190 200 210 220 230 240 250 260 270 280 290 300
80 70 60 50 40 30 20 10 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
1 1 1 1 1 1 1 1 1
1 1 1 1 1 1 1 1 1
100 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 100
100 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 100
100 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 100
100 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 100
100 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 100
100 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 100
100 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 100
100 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 100
100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100 100
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000000 10000000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000000 10000000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000000 10000000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000000 10000000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000000 10000000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 100000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000000 10000000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000000 10000000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000000 10000000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000000 10000000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000000 10000000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000 10000
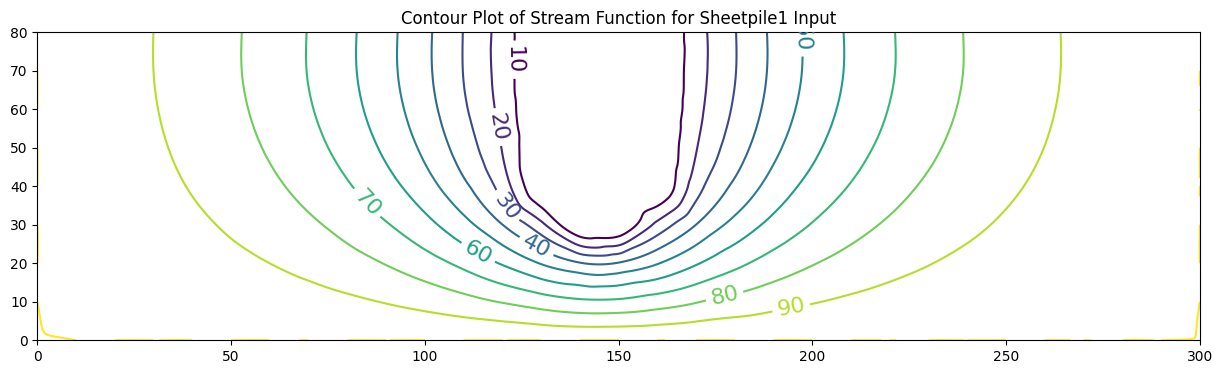
In these examples the value of the stream function is arbitrarily set to range from \(0\) to \(100\). One useful interpretation of stream function values is that their differences indicate the flow fraction (of total flow) that flows between the streamlines (contour lines of constant stream function value).
def sse(matrix1,matrix2):
sse=0.0
nr=len(matrix1) # get row count
nc=len(matrix1[0]) # get column count
for ir in range(nr):
for jc in range(nc):
sse=sse+(matrix1[ir][jc]-matrix2[ir][jc])**2
return(sse)
def update(matrix1,matrix2):
nr=len(matrix1) # get row count
nc=len(matrix1[0]) # get column count
##print(nr,nc)
for ir in range(nr):
for jc in range(nc):
##print(ir,jc)
matrix2[ir][jc]=matrix1[ir][jc]
return(matrix2)
def writearray(matrix):
nr=len(matrix) # get row count
nc=len(matrix[0]) # get column count
import numpy as np
new_list = list(np.around(np.array(matrix), 3))
for ir in range(nr):
print(ir,new_list[ir][:])
return()
localfile = open("StreamFnSheetpile1.txt","r") # connect and read file for 2D gw model
deltax = float(localfile.readline())
deltay = float(localfile.readline())
deltaz = float(localfile.readline())
nrows = int(localfile.readline())
ncols = int(localfile.readline())
tolerance = float(localfile.readline())
maxiter = int(localfile.readline())
distancex = [] # empty list
distancex.append([float(n) for n in localfile.readline().strip().split()])
distancey = [] # empty list
distancey.append([float(n) for n in localfile.readline().strip().split()])
boundarytop = [] #empty list
boundarytop.append([float(n) for n in localfile.readline().strip().split()])
boundarybottom = [] #empty list
boundarybottom.append([int(n) for n in localfile.readline().strip().split()])
boundaryleft = [] #empty list
boundaryleft.append([int(n) for n in localfile.readline().strip().split()])
boundaryright = [] #empty list
boundaryright.append([int(n) for n in localfile.readline().strip().split()])
stream =[] # empty list
for irow in range(nrows):
stream.append([float(n) for n in localfile.readline().strip().split()])
hydcondx = [] # empty list
for irow in range(nrows):
hydcondx.append([float(n) for n in localfile.readline().strip().split()])
hydcondy = [] # empty list
for irow in range(nrows):
hydcondy.append([float(n) for n in localfile.readline().strip().split()])
localfile.close() # Disconnect the file
##
amat = [[0 for j in range(ncols)] for i in range(nrows)]
bmat = [[0 for j in range(ncols)] for i in range(nrows)]
cmat = [[0 for j in range(ncols)] for i in range(nrows)]
dmat = [[0 for j in range(ncols)] for i in range(nrows)]
##
for irow in range(1,nrows-1):
for jcol in range(1,ncols-1):
amat[irow][jcol] = (( hydcondx[irow-1][jcol ]+ hydcondx[irow ][jcol ]) * deltaz ) /(2.0*deltax**2)
bmat[irow][jcol] = (( hydcondx[irow ][jcol ]+ hydcondx[irow+1][jcol ]) * deltaz ) /(2.0*deltax**2)
cmat[irow][jcol] = (( hydcondy[irow ][jcol-1]+ hydcondy[irow ][jcol ]) * deltaz ) /(2.0*deltay**2)
dmat[irow][jcol] = (( hydcondy[irow ][jcol ]+ hydcondy[irow ][jcol+1]) * deltaz ) /(2.0*deltay**2)
##
streamold = [[0 for jc in range(ncols)] for ir in range(nrows)] #force new matrix
streamold = update(stream,streamold) # update
##writearray(stream)
##print("----")
##writearray(streamold)
##print("--------")
tolflag = False
for iter in range(maxiter):
## print("begin iteration")
## writearray(stream)
## print("----")
## writearray(streamold)
## print("--------")
# Boundary Conditions
# first and last row special == no flow boundaries
for jcol in range(ncols):
if boundarytop[0][jcol] == 0: # no - flow at top
stream [0][jcol ] = stream[1][jcol ]
if boundarybottom[0][ jcol ] == 0: # no - flow at bottom
stream [nrows-1][jcol ] = stream[nrows-2][jcol ]
# first and last column special == no flow boundaries
for irow in range(nrows):
if boundaryleft[0][ irow ] == 0:
stream[irow][0] = stream [irow][1] # no - flow at left
if boundaryright[0][ irow ] == 0:
stream [irow][ncols-1] = stream[ irow ][ncols-2] # no - flow at right
# interior updates
for irow in range(1,nrows-1):
for jcol in range(1,ncols-1):
stream[irow][jcol]=( amat[irow][jcol]*stream[irow-1][jcol ]
+bmat[irow][jcol]*stream[irow+1][jcol ]
+cmat[irow][jcol]*stream[irow ][jcol-1]
+dmat[irow][jcol]*stream[irow ][jcol+1])/(amat[ irow][jcol ]+ bmat[ irow][jcol ]+ cmat[ irow][jcol ]+ dmat[ irow][jcol ])
# test for stopping iterations
## print("end iteration")
## writearray(stream)
## print("----")
## writearray(streamold)
percentdiff = sse(stream,streamold)
if percentdiff <= tolerance:
print("Exit iterations in Stream Function because tolerance met ")
print("Iterations =" , iter+1 ) ;
tolflag = True
break
streamold = update(stream,streamold)
print("End Calculations")
print("Iterations = ",iter+1)
print("Closure Error = ",round(percentdiff,3))
print("Stream Function Map")
print("----")
writearray(stream)
print("----")
####################### Contour Plot ###############
my_xyz2 = [] # empty list
for irow in range(nrows):
for jcol in range(ncols):
my_xyz2.append([distancex[0][jcol],distancey[0][irow],stream[irow][jcol]])
import pandas
my_xyz2 = pandas.DataFrame(my_xyz2) # convert into a data frame
#print(my_xyz) # activate to examine the dataframe
import numpy
import matplotlib.pyplot
from scipy.interpolate import griddata
# extract lists from the dataframe
coord_x2 = my_xyz2[0].values.tolist() # column 0 of dataframe
coord_y2 = my_xyz2[1].values.tolist() # column 1 of dataframe
coord_z2 = my_xyz2[2].values.tolist() # column 2 of dataframe
coord_xy2 = numpy.column_stack((coord_x2, coord_y2))
# Set plotting range in original data units
lon = numpy.linspace(min(coord_x2), max(coord_x2), 3000)
lat = numpy.linspace(min(coord_y2), max(coord_y2), 800)
X2, Y2 = numpy.meshgrid(lon, lat)
# Grid the data; use linear interpolation (choices are nearest, linear, cubic)
Z2 = griddata(numpy.array(coord_xy2), numpy.array(coord_z2), (X2, Y2), method='cubic')
# Build the map
fig, ax = matplotlib.pyplot.subplots()
fig.set_size_inches(15, 4)
levels = [10,20,30,40,50,60,70,80,90,100]
CS2 = ax.contour(X2, Y2, Z2, levels )
ax.clabel(CS2, inline=2, fontsize=16)
ax.set_title('Contour Plot of Stream Function for Sheetpile1 Input')
Exit iterations in Stream Function because tolerance met
Iterations = 396
End Calculations
Iterations = 396
Closure Error = 0.0
Stream Function Map
----
0 [100. 96.845 93.555 89.987 85.989 81.394 76.015 69.642 62.046
52.99 42.26 29.74 15.542 0.191 0. 0. 0.19 15.469
29.592 42.031 52.669 61.62 69.093 75.318 80.519 84.898 88.632
91.878 94.773 97.442 100. ]
1 [100. 96.845 93.555 89.987 85.989 81.394 76.015 69.642 62.046
52.99 42.26 29.74 15.542 0.191 0.16 0.16 0.19 15.469
29.592 42.031 52.669 61.62 69.093 75.318 80.519 84.898 88.632
91.878 94.773 97.442 100. ]
2 [100. 96.982 93.832 90.416 86.586 82.178 77.008 70.865 63.507
54.663 44.05 31.419 16.694 0.337 0.304 0.304 0.336 16.625
31.277 43.83 54.356 63.099 70.34 76.342 81.342 85.543 89.121
92.229 94.999 97.553 100. ]
3 [100. 97.249 94.377 91.26 87.76 83.724 78.976 73.304 66.452
58.105 47.859 35.19 19.48 0.47 0.432 0.432 0.47 19.416
35.061 47.658 57.826 66.081 72.825 78.369 82.963 86.81 90.08
92.916 95.443 97.77 100. ]
4 [100. 97.636 95.168 92.486 89.47 85.984 81.867 76.921 70.894
63.447 54.089 42.004 25.566 0.634 0.541 0.541 0.634 25.509
41.891 53.916 63.207 70.575 76.51 81.346 85.33 88.655 91.473
93.914 96.085 98.085 100. ]
5 [100. 98.128 96.172 94.045 91.65 88.875 85.587 81.619 76.754
70.701 63.045 53.171 40.146 22.721 0.665 0.665 22.703 40.095
53.079 62.907 70.511 76.502 81.294 85.175 88.358 91.005 93.244
95.18 96.901 98.484 100. ]
6 [100. 98.703 97.348 95.872 94.209 92.279 89.987 87.213 83.804
79.559 74.22 67.487 59.128 49.437 40.688 40.683 49.419 59.09
67.422 74.124 79.426 83.629 86.988 89.702 91.921 93.763 95.318
96.661 97.853 98.95 100. ]
7 [100. 99.336 98.643 97.888 97.036 96.046 94.869 93.443 91.689
89.509 86.79 83.43 79.442 75.211 71.965 71.962 75.201 79.422
83.396 86.74 89.442 91.6 93.328 94.723 95.863 96.808 97.604
98.292 98.902 99.463 100. ]
8 [100. 100. 100. 100. 100. 100. 100. 100. 100. 100. 100. 100. 100. 100.
100. 100. 100. 100. 100. 100. 100. 100. 100. 100. 100. 100. 100. 100.
100. 100. 100.]
----
Text(0.5, 1.0, 'Contour Plot of Stream Function for Sheetpile1 Input')
Now both plots on same canvass
# Head (Velocity Potential Map)
# Grid the data; use linear interpolation (choices are nearest, linear, cubic)
Z = griddata(numpy.array(coord_xy), numpy.array(coord_z), (X, Y), method='cubic')
Z2 = griddata(numpy.array(coord_xy2), numpy.array(coord_z2), (X2, Y2), method='cubic')
# Build the map
fig, ax = matplotlib.pyplot.subplots()
fig.set_size_inches(15, 4)
levels1 = [1,2,3,4,5,6,7,8,9]
CS = ax.contour(X, Y, Z, levels1)
levels2 = [10,20,30,40,50,60,70,80,90]
CS2 = ax.contour(X2, Y2, Z2, levels2 )
ax.clabel(CS, inline=2, fontsize=12)
ax.clabel(CS2, inline=2, fontsize=12)
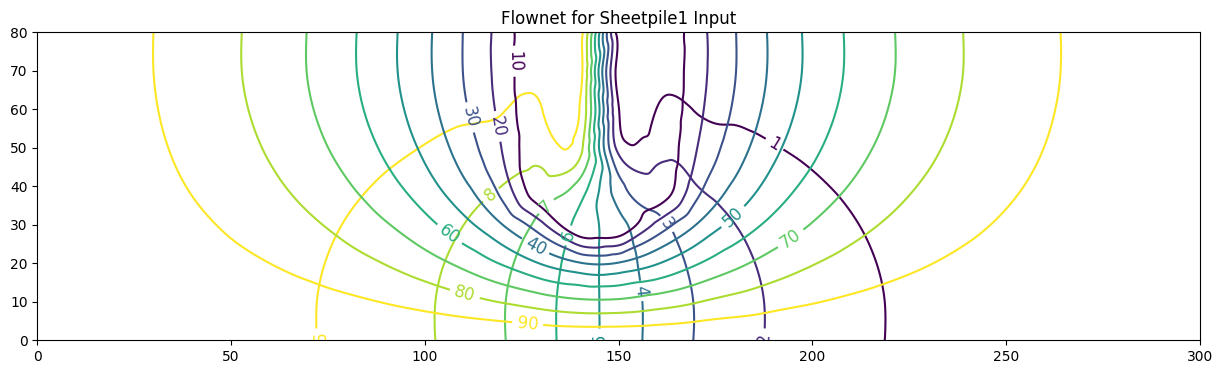
ax.set_title('Flownet for Sheetpile1 Input')
Text(0.5, 1.0, 'Flownet for Sheetpile1 Input')
Tip
Using AI to improve your (already working) code: a practical framework Goal: keep your code correct, make it clearer/faster/safer—and learn something in the process.
Set the ground rules (before you paste code)
Snapshot a baseline: save the working script + sample inputs/outputs.
State objectives: e.g., “modularize,” “add tests,” “speed up by 2×,” “remove globals.”
List invariants: what must not change (I/O format, algorithm, results within tolerance).
Share minimal context: problem description, performance pain points, constraints (no new deps, Py≥3.10, runs headless, etc.).
Provide a small, runnable example: tiny input + expected output for quick checks.
Ask for a plan, then patches
First request: “Propose a refactor plan with milestones (interfaces, tests, perf check).”
Work in small steps (e.g., extract functions → add dataclasses → add tests → add CLI).
Prefer diffs or focused snippets over full rewrites to avoid regressions.
Build safety nets
Tests: ask for unit tests (edge cases + typical), fixtures, and a quick benchmark harness.
Determinism: specify seeds and tolerances (e.g., atol=1e-6), and record environment info.
Validation hooks: print/plot key diagnostics to confirm physics/logic after each change.
Check the work
Require a short rationale for each change (why it helps; complexity trade-offs).
Watch for red flags: invented APIs, silent dependency creep, algorithm swaps, huge boilerplate.
Inspect performance: time/memory before vs after; accept only measurable wins.
Document and package
Ask for docstrings, type hints, and a concise README (usage, assumptions, limits).
Ensure error messages guide the user; add logging where it helps debugging.
Keep your license/copyright headers intact.
Prompt templates (copy/paste)
“Refactor into a module with read_input, assemble_coeffs, solve, plot. Keep I/O identical.”
“Add dataclasses for grid/boundaries; no new dependencies; include type hints.”
“Write pytest tests for 3 cases (baseline, edge, heterogeneity); set numeric tolerances.”
“Suggest a safe SOR acceleration (ω selection), show where it slots in, and guard with a flag.”
“Produce a minimal benchmark script and report speedup on the provided sample.”
Remember: AI is a pair programmer, not an oracle. Keep runs tight, verify often, and merge improvements only when they pass your tests and preserve the science.
What follows below are two modules saved as .py files which are loaded in an identical fashion as a library or any other python module. As you shall observe they make the model build/display much more tidy. They implement the algorithm identically, but are modular.
I supplied the code above with the following prompt:
Attached is my potential flow code. Can you refactor into logical modules, and enforce best programming practices. As this is pedagogical - error checking is not vital, but elegant fails are desirable.
The results are two modules potential_flow_mod.py and flownet_plotter.py
potential_flow_mod.py
# potential_flow_mod.py
#
# Built collaboratively by Sensei and OpenAI ChatGPT on 2025-08-14;
# Derived from the classroom “PotentialFnSheetpile1” script.
# Gauss–Seidel iterative solver for Laplacian-type linear systems (in-place updates)
#
from dataclasses import dataclass
from typing import Tuple
import numpy as np
import matplotlib.pyplot as plt
from scipy.interpolate import griddata
@dataclass
class GridSpec:
deltax: float
deltay: float
deltaz: float
nrows: int
ncols: int
distancex: np.ndarray
distancey: np.ndarray
@dataclass
class Boundaries:
top: np.ndarray
bottom: np.ndarray
left: np.ndarray
right: np.ndarray
@dataclass
class ModelData:
grid: GridSpec
bounds: Boundaries
head0: np.ndarray
Kx: np.ndarray
Ky: np.ndarray
tolerance: float
maxiter: int
def read_potential_input(path: str) -> ModelData:
with open(path, "r") as f:
deltax = float(f.readline().strip())
deltay = float(f.readline().strip())
deltaz = float(f.readline().strip())
nrows = int(f.readline().strip())
ncols = int(f.readline().strip())
tolerance = float(f.readline().strip())
maxiter = int(f.readline().strip())
distancex = np.array([float(x) for x in f.readline().strip().split()], dtype=float)
distancey = np.array([float(y) for y in f.readline().strip().split()], dtype=float)
boundarytop = np.array([float(v) for v in f.readline().strip().split()], dtype=float)
boundarybottom = np.array([int(v) for v in f.readline().strip().split()], dtype=int)
boundaryleft = np.array([int(v) for v in f.readline().strip().split()], dtype=int)
boundaryright = np.array([int(v) for v in f.readline().strip().split()], dtype=int)
head0 = np.zeros((nrows, ncols), dtype=float)
for i in range(nrows):
head0[i, :] = np.array([float(v) for v in f.readline().strip().split()], dtype=float)
Kx = np.zeros((nrows, ncols), dtype=float)
for i in range(nrows):
Kx[i, :] = np.array([float(v) for v in f.readline().strip().split()], dtype=float)
Ky = np.zeros((nrows, ncols), dtype=float)
for i in range(nrows):
Ky[i, :] = np.array([float(v) for v in f.readline().strip().split()], dtype=float)
grid = GridSpec(deltax, deltay, deltaz, nrows, ncols, distancex, distancey)
bounds = Boundaries(boundarytop, boundarybottom, boundaryleft, boundaryright)
return ModelData(grid, bounds, head0, Kx, Ky, tolerance, maxiter)
def assemble_coefficients(Kx: np.ndarray, Ky: np.ndarray, grid: GridSpec):
nr, nc = grid.nrows, grid.ncols
A = np.zeros((nr, nc), dtype=float)
B = np.zeros((nr, nc), dtype=float)
C = np.zeros((nr, nc), dtype=float)
D = np.zeros((nr, nc), dtype=float)
dx2 = grid.deltax ** 2
dy2 = grid.deltay ** 2
dz = grid.deltaz
for i in range(1, nr - 1):
for j in range(1, nc - 1):
A[i, j] = ((Kx[i - 1, j] + Kx[i, j]) * dz) / (2.0 * dx2)
B[i, j] = ((Kx[i, j] + Kx[i + 1, j]) * dz) / (2.0 * dx2)
C[i, j] = ((Ky[i, j - 1] + Ky[i, j]) * dz) / (2.0 * dy2)
D[i, j] = ((Ky[i, j] + Ky[i, j + 1]) * dz) / (2.0 * dy2)
return A, B, C, D
def apply_no_flow_edges(head: np.ndarray, bounds: Boundaries) -> None:
nr, nc = head.shape
for j in range(nc):
if bounds.top[j] == 0:
head[0, j] = head[1, j]
if bounds.bottom[j] == 0:
head[nr - 1, j] = head[nr - 2, j]
for i in range(nr):
if bounds.left[i] == 0:
head[i, 0] = head[i, 1]
if bounds.right[i] == 0:
head[i, nc - 1] = head[i, nc - 2]
def gauss_seidel_sweep(head: np.ndarray, A: np.ndarray, B: np.ndarray, C: np.ndarray, D: np.ndarray) -> None:
nr, nc = head.shape
for i in range(1, nr - 1):
for j in range(1, nc - 1):
wsum = A[i, j] + B[i, j] + C[i, j] + D[i, j]
if wsum > 0.0:
head[i, j] = (
A[i, j] * head[i - 1, j] +
B[i, j] * head[i + 1, j] +
C[i, j] * head[i, j - 1] +
D[i, j] * head[i, j + 1]
) / wsum
def solve_potential(model, verbose: bool = True):
grid, bounds = model.grid, model.bounds
head = model.head0.copy()
head_old = head.copy()
A, B, C, D = assemble_coefficients(model.Kx, model.Ky, grid)
tol = model.tolerance
maxiter = model.maxiter
for it in range(1, maxiter + 1):
apply_no_flow_edges(head, bounds)
gauss_seidel_sweep(head, A, B, C, D)
sse = float(np.sum((head - head_old) ** 2))
if verbose and (it % 50 == 0 or it == 1):
print(f"Iter {it:6d} | SSE={sse:.6e}")
if sse <= tol:
if verbose:
print("Exit iterations in velocity potential because tolerance met")
print("Iterations =", it)
return head, it, sse
head_old[:, :] = head
if verbose:
print("Reached max iterations.")
return head, maxiter, float(np.sum((head - head_old) ** 2))
def plot_head_contours(distancex: np.ndarray, distancey: np.ndarray, head: np.ndarray,
title: str = "Contour Plot of Heads", nx: int = 1200, ny: int = 600, method: str = "cubic"):
Xpts, Ypts = np.meshgrid(distancex, distancey)
pts = np.column_stack([Xpts.ravel(), Ypts.ravel()])
vals = head.ravel()
gx = np.linspace(distancex.min(), distancex.max(), nx)
gy = np.linspace(distancey.min(), distancey.max(), ny)
GX, GY = np.meshgrid(gx, gy)
GZ = griddata(pts, vals, (GX, GY), method=method)
fig, ax = plt.subplots(figsize=(12, 4))
CS = ax.contour(GX, GY, GZ, levels=10)
ax.clabel(CS, inline=2, fontsize=12)
ax.set_title(title)
ax.set_xlabel("x (distance units)")
ax.set_ylabel("y (distance units)")
fig.tight_layout()
return fig
flownet_plotter.py
# flownet_plotter.py
#
# Built collaboratively by Sensei and OpenAI ChatGPT on 2025-08-14.
# Overlays equipotential (head) and stream-function contours on a common grid to form a flownet.
# Reads two 3-column XYZ files (x y value), grids via scipy.griddata, and plots head (solid) + stream (dashed).
# Includes a helper to write XYZ from regular grids and a CLI for quick use; no explicit colors are set.
#
import numpy as np
import matplotlib.pyplot as plt
from scipy.interpolate import griddata
from matplotlib.lines import Line2D
import argparse
import os
def read_xyz(path):
"""
Read an XYZ file with 3 columns: x, y, value.
Delimiters may be commas or whitespace.
Lines starting with '#' or '//' are ignored.
Returns: (x, y, z) as 1D numpy arrays.
"""
xs, ys, zs = [], [], []
with open(path, 'r', encoding='utf-8') as f:
for line in f:
s = line.strip()
if not s:
continue
if s.startswith('#') or s.startswith('//'):
continue
s = s.replace(',', ' ')
parts = s.split()
if len(parts) < 3:
continue
try:
x, y, z = float(parts[0]), float(parts[1]), float(parts[2])
except ValueError:
continue
xs.append(x); ys.append(y); zs.append(z)
if len(xs) == 0:
raise ValueError(f"No valid data found in {path}")
return np.asarray(xs), np.asarray(ys), np.asarray(zs)
def grid_xyz(x, y, z, nx=1200, ny=600, extent=None, method='cubic'):
"""
Grid scattered (x,y,z) onto a regular mesh using scipy.griddata.
extent: (xmin, xmax, ymin, ymax); if None, computed from data.
Returns: GX, GY, GZ
"""
if extent is None:
xmin, xmax = np.min(x), np.max(x)
ymin, ymax = np.min(y), np.max(y)
else:
xmin, xmax, ymin, ymax = extent
gx = np.linspace(xmin, xmax, nx)
gy = np.linspace(ymin, ymax, ny)
GX, GY = np.meshgrid(gx, gy)
pts = np.column_stack([x, y])
GZ = griddata(pts, z, (GX, GY), method=method)
return GX, GY, GZ
"""def write_xyz_grid(distancex, distancey, array2d, path):
"""
# Helper to write a regular grid (distancex, distancey, array) into
# 3-column XYZ format. Each row: x y value
"""
X, Y = np.meshgrid(distancex, distancey)
xyz = np.column_stack([X.ravel(), Y.ravel(), array2d.ravel()])
print("in write xyz")
np.savetxt(
path,
xyz,
fmt="%.6g",
delimiter=" ",
header="x y value", # do NOT include '\n' here
comments="# " # keeps a normal '# ' header line
)
with open(path, 'w', encoding='utf-8') as f:
f.write('# x y value')
for i, yy in enumerate(distancey):
for j, xx in enumerate(distancex):
f.write(f"{xx} {yy} {array2d[i, j]}"+"\n")
"""
def write_xyz_grid(distancex, distancey, array2d, path,
label="value", precision=6, transpose=False,
newline="\n", verbose=False):
"""
Write a regular grid to 3-column XYZ text: x y <label>
- distancex: (ncols,)
- distancey: (nrows,)
- array2d: (nrows, ncols)
- transpose=True if your array orientation needs flipping
"""
X, Y = np.meshgrid(distancex, distancey)
Z = array2d.T if transpose else array2d
xyz = np.column_stack([X.ravel(), Y.ravel(), Z.ravel()])
np.savetxt(
path,
xyz,
fmt=f"%.{precision}g",
delimiter=" ",
header=f"x y {label}", # reader ignores '#' lines
comments="# ",
newline=newline # ensures real newlines on all OSes
)
if verbose:
print(f"Wrote {xyz.shape[0]} rows to {path}")
def plot_flownet(head_file, stream_file,
head_levels=None, stream_levels=None,
nx=1200, ny=600, method='cubic',
title='Flownet', figsize=(7.5, 4),
head_label_size=12, stream_label_size=10,
save_png=None, transparent=True):
"""
Plot head and stream-function contours from two XYZ files on a common grid.
Parameters
----------
head_file : str
Path to XYZ file with (x, y, head).
stream_file : str
Path to XYZ file with (x, y, stream function).
head_levels : list[float] or None
Contour levels for head. If None, auto-selected.
stream_levels : list[float] or None
Contour levels for stream function. If None, auto-selected.
nx, ny : int
Grid size for interpolation.
method : str
'nearest', 'linear', or 'cubic' interpolation.
title : str
Figure title.
figsize : tuple
Figure size in inches.
head_label_size, stream_label_size : int
Font sizes for contour labels.
save_png : str or None
If provided, save to this path.
transparent : bool
Save PNG with transparent background.
Returns
-------
fig : matplotlib.figure.Figure
ax : matplotlib.axes.Axes
"""
x1, y1, z1 = read_xyz(head_file)
x2, y2, z2 = read_xyz(stream_file)
# Common extent from union of both datasets
xmin = min(np.min(x1), np.min(x2))
xmax = max(np.max(x1), np.max(x2))
ymin = min(np.min(y1), np.min(y2))
ymax = max(np.max(y1), np.max(y2))
extent = (xmin, xmax, ymin, ymax)
# Grid both to the same mesh
GX, GY, GZ1 = grid_xyz(x1, y1, z1, nx=nx, ny=ny, extent=extent, method=method)
_, _, GZ2 = grid_xyz(x2, y2, z2, nx=nx, ny=ny, extent=extent, method=method)
# Auto levels if not provided
def auto_levels(arr, n=10):
finite = np.asarray(arr[np.isfinite(arr)])
if finite.size == 0:
return None
lo, hi = np.percentile(finite, [10, 90]) if np.ptp(finite) > 0 else (finite.min(), finite.max())
if hi == lo:
hi = lo + 1.0 # avoid degenerate contour
return np.linspace(lo, hi, n)
if head_levels is None:
head_levels = auto_levels(GZ1, n=8)
if stream_levels is None:
stream_levels = auto_levels(GZ2, n=10)
fig, ax = plt.subplots(figsize=figsize)
# Head: solid lines
CS1 = ax.contour(GX, GY, GZ1, levels=head_levels)
ax.clabel(CS1, inline=2, fontsize=head_label_size)
# Stream function: dashed lines (no explicit color set)
CS2 = ax.contour(GX, GY, GZ2, levels=stream_levels, linestyles='--')
ax.clabel(CS2, inline=2, fontsize=stream_label_size)
ax.set_title(title)
ax.set_xlabel('x')
ax.set_ylabel('y')
# Legend using proxy artists (default line color, different linestyles)
proxies = [
Line2D([0], [0]),
Line2D([0], [0], linestyle='--')
]
ax.legend(proxies, ['Potential function (head)', 'Stream function (streamlines)'], loc='best', frameon=True)
fig.tight_layout()
if save_png:
fig.savefig(save_png, dpi=300, bbox_inches='tight', transparent=transparent)
return fig, ax
def _parse_levels(s):
"""Parse a comma-separated string of numbers into a list of floats."""
if s is None:
return None
s = s.strip()
if not s:
return None
return [float(x) for x in s.split(',')]
def main():
parser = argparse.ArgumentParser(description='Flownet plotter: overlay head and stream function contours.')
parser.add_argument('head_file', help='Path to head XYZ file (x y head).')
parser.add_argument('stream_file', help='Path to stream-function XYZ file (x y psi).')
parser.add_argument('--levels-head', type=str, default=None, help='Comma-separated head contour levels, e.g., "5,6,7,8".')
parser.add_argument('--levels-stream', type=str, default=None, help='Comma-separated stream contour levels, e.g., "10,20,30".')
parser.add_argument('--nx', type=int, default=1200, help='Interpolation grid size in x.')
parser.add_argument('--ny', type=int, default=600, help='Interpolation grid size in y.')
parser.add_argument('--method', type=str, default='cubic', choices=['nearest','linear','cubic'], help='griddata method.')
parser.add_argument('--title', type=str, default='Flownet', help='Figure title.')
parser.add_argument('--save', type=str, default=None, help='Output PNG path.')
parser.add_argument('--transparent', action='store_true', help='Save with transparent background.')
args = parser.parse_args()
fig, ax = plot_flownet(
args.head_file, args.stream_file,
head_levels=_parse_levels(args.levels_head),
stream_levels=_parse_levels(args.levels_stream),
nx=args.nx, ny=args.ny, method=args.method,
title=args.title, save_png=args.save, transparent=args.transparent
)
plt.show()
if __name__ == '__main__':
main()
Now apply to the same input files
%reset -f
from potential_flow_mod import read_potential_input, solve_potential
from flownet_plotter import write_xyz_grid, plot_flownet
# 1) Solve velocity potential (head)
modelp = read_potential_input("PotentialFnSheetpile1.txt")
head, niter, closure = solve_potential(modelp, verbose=False)
#fig = plot_head_contours(modelp.grid.distancex, modelp.grid.distancey, head, title="Contour Plot of Heads from Sheetpile1 Input")
# 2) Write solver outputs as XYZ (3 columns: x y value)
write_xyz_grid(modelp.grid.distancex, modelp.grid.distancey, head, "head_xyz.txt")
# 3) Solve stream function (streamlines)
models = read_potential_input("StreamFnSheetpile1.txt")
stream, niter, closure = solve_potential(models, verbose=False)
#fig = plot_head_contours(models.grid.distancex, models.grid.distancey, stream, title="Contour Plot of Heads from Sheetpile1 Input")
# 4) Write solver outputs as XYZ (3 columns: x y value)
write_xyz_grid(models.grid.distancex, models.grid.distancey, stream, "stream_xyz.txt")
# 5) Make the flownet
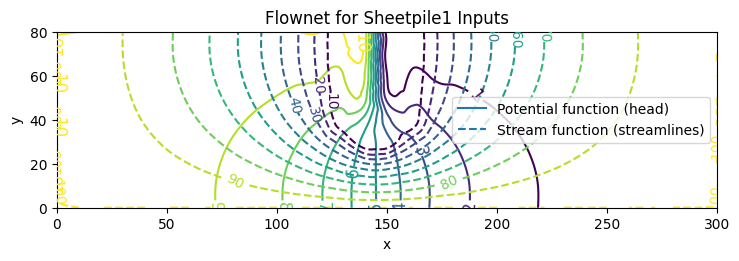
fig, ax = plot_flownet(
"head_xyz.txt", "stream_xyz.txt",
head_levels=[1,2,3,4,5,6,7,8,9,10],
stream_levels=[10,20,30,40,50,60,70,80,90,100],
title="Flownet for Sheetpile1 Inputs",
save_png="flownet1.png",
transparent=True
)
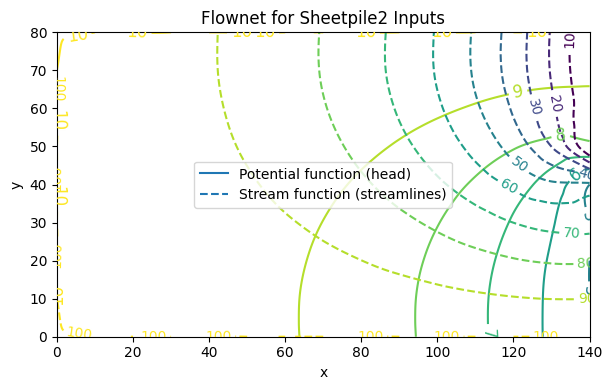
Flownet Example 2 : Pore Pressure under a Dam (Domain Decomposition approach)#
The second way to solve this example, and perhaps better is to take advantage of the symmetry and cut the domain in half, as
In this method we can actually specify the sheetpile as a boundary, and we will obtain the about same results, but only need to supply half as much input data.
Here’s the input file for the half-domain head calculations
10
10
1
9
15
1e-16
5000
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140
80 70 60 50 40 30 20 10 0
1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1 1 1 1 1 1 1 1 1
0 0 0 0 1 1 1 1 1
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10
10 10 10 10 10 10 10 10 10 10 10 10 10 10 10
10 10 10 10 10 10 10 10 10 10 10 10 10 10 5
10 10 10 10 10 10 10 10 10 10 10 10 10 10 5
10 10 10 10 10 10 10 10 10 10 10 10 10 10 5
10 10 10 10 10 10 10 10 10 10 10 10 10 10 5
10 10 10 10 10 10 10 10 10 10 10 10 10 10 5
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001 0.0001
%reset -f
from potential_flow_mod import read_potential_input, solve_potential
from flownet_plotter import write_xyz_grid, plot_flownet
# 1) Solve velocity potential (head)
modelp = read_potential_input("PotentialFnSheetpile2.txt")
head, niter, closure = solve_potential(modelp, verbose=False)
#fig = plot_head_contours(modelp.grid.distancex, modelp.grid.distancey, head, title="Contour Plot of Heads from Sheetpile1 Input")
# 2) Write solver outputs as XYZ (3 columns: x y value)
write_xyz_grid(modelp.grid.distancex, modelp.grid.distancey, head, "head_xyz.txt")
# 3) Solve stream function (streamlines)
models = read_potential_input("StreamFnSheetpile2.txt")
stream, niter, closure = solve_potential(models, verbose=False)
#fig = plot_head_contours(models.grid.distancex, models.grid.distancey, stream, title="Contour Plot of Heads from Sheetpile1 Input")
# 4) Write solver outputs as XYZ (3 columns: x y value)
write_xyz_grid(models.grid.distancex, models.grid.distancey, stream, "stream_xyz.txt")
# 5) Make the flownet
fig, ax = plot_flownet(
"head_xyz.txt", "stream_xyz.txt",
head_levels=[1,2,3,4,5,6,7,8,9,10],
stream_levels=[10,20,30,40,50,60,70,80,90,100],
title="Flownet for Sheetpile2 Inputs",
save_png="flownet2.png",
transparent=True
)
Summary#
This chapter refreshed the governing equations of groundwater flow and the advection–dispersion equation for transport, emphasizing how Darcy’s law and mass conservation underpin both. Because you have seen these before, we focused on the essentials: storage terms (specific yield vs. specific storage), anisotropic conductivities, and how velocity fields from the flow solution drive transport. The aim was not to re-derive everything in full, but to recall the physics you need at your fingertips when you start building and checking models.
We then turned to flownets as a practical, low-overhead way to explore boundary effects and heterogeneity. Using a homebrew potential solver and flownet plotter, we showed how equipotential lines and stream functions can be overlaid to visualize flow patterns, estimate gradients, and even approximate capture zones—all without booting up MODFLOW. This approach is ideal for rapid “what-if” tests, classroom demonstrations, and early scoping on real sites. We noted anisotropy conceptually (directional conductivities) and left its detailed illustration as an exercise; extending the flownet to anisotropic cases is a natural next step. Together, these tools bridge theory and practice: they keep the physics honest while giving you fast, reproducible visuals that support interpretation and decision-making.