Poison Mushrooms¶
An application example of multiple logistic regression.
Here we will have to encode the categories, then build the model.
attributes=['poison-class','cap-shape','cap-surface','cap-color','bruises','odor','gill-attachment','gill-spacing','gill-size','gill-color','stalk-shape','stalk-root','stalk-surface-above-ring','stalk-surface-below-ring','stalk-color-above-ring','stalk-color-below-ring','veil-type','veil-color','ring-number','ring-type','spore-print-color','population','habitat']
len(attributes)
23
import sys # Module to process commands to/from the OS using a shell-type syntax
import requests
remote_url="http://54.243.252.9/ce-5319-webroot/ce5319jb/lessons/logisticregression/agaricus-lepiota.data" # set the url
rget = requests.get(remote_url, allow_redirects=True) # get the remote resource, follow imbedded links
localfile = open('poisonmushroom.csv','wb') # open connection to a local file same name as remote
localfile.write(rget.content) # extract from the remote the contents,insert into the local file same name
localfile.close() # close connection to the local file
# delete file if it exists
import pandas as pd
mymushroom = pd.read_csv('poisonmushroom.csv',header=None)
req_col_names = attributes
curr_col_names = list(mymushroom.columns)
mapper = {}
for i, name in enumerate(curr_col_names):
mapper[name] = req_col_names[i]
mymushroom = mymushroom.rename(columns=mapper)
interim = pd.DataFrame(mymushroom)
mymushroom.head(20)
| poison-class | cap-shape | cap-surface | cap-color | bruises | odor | gill-attachment | gill-spacing | gill-size | gill-color | ... | stalk-surface-below-ring | stalk-color-above-ring | stalk-color-below-ring | veil-type | veil-color | ring-number | ring-type | spore-print-color | population | habitat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | p | x | s | n | t | p | f | c | n | k | ... | s | w | w | p | w | o | p | k | s | u |
| 1 | e | x | s | y | t | a | f | c | b | k | ... | s | w | w | p | w | o | p | n | n | g |
| 2 | e | b | s | w | t | l | f | c | b | n | ... | s | w | w | p | w | o | p | n | n | m |
| 3 | p | x | y | w | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | k | s | u |
| 4 | e | x | s | g | f | n | f | w | b | k | ... | s | w | w | p | w | o | e | n | a | g |
| 5 | e | x | y | y | t | a | f | c | b | n | ... | s | w | w | p | w | o | p | k | n | g |
| 6 | e | b | s | w | t | a | f | c | b | g | ... | s | w | w | p | w | o | p | k | n | m |
| 7 | e | b | y | w | t | l | f | c | b | n | ... | s | w | w | p | w | o | p | n | s | m |
| 8 | p | x | y | w | t | p | f | c | n | p | ... | s | w | w | p | w | o | p | k | v | g |
| 9 | e | b | s | y | t | a | f | c | b | g | ... | s | w | w | p | w | o | p | k | s | m |
| 10 | e | x | y | y | t | l | f | c | b | g | ... | s | w | w | p | w | o | p | n | n | g |
| 11 | e | x | y | y | t | a | f | c | b | n | ... | s | w | w | p | w | o | p | k | s | m |
| 12 | e | b | s | y | t | a | f | c | b | w | ... | s | w | w | p | w | o | p | n | s | g |
| 13 | p | x | y | w | t | p | f | c | n | k | ... | s | w | w | p | w | o | p | n | v | u |
| 14 | e | x | f | n | f | n | f | w | b | n | ... | f | w | w | p | w | o | e | k | a | g |
| 15 | e | s | f | g | f | n | f | c | n | k | ... | s | w | w | p | w | o | p | n | y | u |
| 16 | e | f | f | w | f | n | f | w | b | k | ... | s | w | w | p | w | o | e | n | a | g |
| 17 | p | x | s | n | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | k | s | g |
| 18 | p | x | y | w | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | n | s | u |
| 19 | p | x | s | n | t | p | f | c | n | k | ... | s | w | w | p | w | o | p | n | s | u |
20 rows × 23 columns
mymushroom.head(10)
| poison-class | cap-shape | cap-surface | cap-color | bruises | odor | gill-attachment | gill-spacing | gill-size | gill-color | ... | stalk-surface-below-ring | stalk-color-above-ring | stalk-color-below-ring | veil-type | veil-color | ring-number | ring-type | spore-print-color | population | habitat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | p | x | s | n | t | p | f | c | n | k | ... | s | w | w | p | w | o | p | k | s | u |
| 1 | e | x | s | y | t | a | f | c | b | k | ... | s | w | w | p | w | o | p | n | n | g |
| 2 | e | b | s | w | t | l | f | c | b | n | ... | s | w | w | p | w | o | p | n | n | m |
| 3 | p | x | y | w | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | k | s | u |
| 4 | e | x | s | g | f | n | f | w | b | k | ... | s | w | w | p | w | o | e | n | a | g |
| 5 | e | x | y | y | t | a | f | c | b | n | ... | s | w | w | p | w | o | p | k | n | g |
| 6 | e | b | s | w | t | a | f | c | b | g | ... | s | w | w | p | w | o | p | k | n | m |
| 7 | e | b | y | w | t | l | f | c | b | n | ... | s | w | w | p | w | o | p | n | s | m |
| 8 | p | x | y | w | t | p | f | c | n | p | ... | s | w | w | p | w | o | p | k | v | g |
| 9 | e | b | s | y | t | a | f | c | b | g | ... | s | w | w | p | w | o | p | k | s | m |
10 rows × 23 columns
def p0(stringvalue):
if stringvalue == 'e':
p0 = 0
elif stringvalue == 'p':
p0 = 1
else:
raise Exception("Encoding failed in p0 missing data maybe?")
return(p0)
######################################################################
# Feature Encoding Functions using a Simple Substitution Cipher ##
######################################################################
def c1(stringvalue):
#cap-shape: bell=b,conical=c,convex=x,flat=f,knobbed=k,sunken=s
ncode=True # set exception flag
alphabet=['b','c','x','f','k','s']
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c1=i
ncode=False # if encoding swithc flag value
if ncode:
raise Exception("Encoding failed in c1 missing data maybe?")
return(c1)
######################################################################
def c2(stringvalue):
#cap-surface: fibrous=f,grooves=g,scaly=y,smooth=s
ncode=True
alphabet=['f','g','y','s']
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c2=i
ncode=False
if ncode:
raise Exception("Encoding failed in c2 missing data maybe?")
return(c2)
######################################################################
def c3(stringvalue):
#cap-color: brown=n,buff=b,cinnamon=c,gray=g,green=r,pink=p,purple=u,red=e,white=w,yellow=y
ncode=True
alphabet=['n','b','c','g','r','p','u','e','w','y']
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c3=i
ncode=False
if ncode:
raise Exception("Encoding failed in c3 missing data maybe?")
return(c3)
######################################################################
def c4(stringvalue): #this is a simple binary encoding column
#bruises?:bruises=t,no=f
ncode=True
alphabet=['f','t']
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c4=i
ncode=False
if ncode:
raise Exception("Encoding failed in c4 missing data maybe?")
return(c4)
######################################################################
def c5(stringvalue):
#odor: almond=a,anise=l,creosote=c,fishy=y,foul=f,musty=m,none=n,pungent=p,spicy=s
ncode=True
alphabet=['a','l','c','y','f','m','n','p','s']
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c5=i
ncode=False
if ncode:
raise Exception("Encoding failed in c5 missing data maybe?")
return(c5)
######################################################################
def c6(stringvalue):
#gill-attachment: attached=a,descending=d,free=f,notched=n
ncode=True
alphabet=['a','d','f','n']
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c6=i
ncode=False
if ncode:
raise Exception("Encoding failed in c6 missing data maybe?")
return(c6)
######################################################################
def c7(stringvalue):
#gill-spacing: close=c,crowded=w,distant=d
ncode=True
alphabet=['c','w','d']
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c7=i
ncode=False
if ncode:
raise Exception("Encoding failed in c7 missing data maybe?")
return(c7)
######################################################################
def c8(stringvalue):
#gill-size: broad=b,narrow=n
ncode=True
alphabet=['b','n']
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c8=i
ncode=False
if ncode:
raise Exception("Encoding failed in c8 missing data maybe?")
return(c8)
######################################################################
def c9(stringvalue):
#gill-color: black=k,brown=n,buff=b,chocolate=h,gray=g,green=r,orange=o,pink=p,purple=u,red=e,white=w,yellow=y
ncode=True
alphabet=['k','n','b','h','g','r','o','p','u','e','w','y']
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c9=i
ncode=False
if ncode:
raise Exception("Encoding failed in c9 missing data maybe?")
return(c9)
######################################################################
def c10(stringvalue):
#stalk-shape:enlarging=e,tapering=t
ncode=True
alphabet=['e','t']
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c10=i
ncode=False
if ncode:
raise Exception("Encoding failed in c10 missing data maybe?")
return(c10)
######################################################################
def c11(stringvalue):
#stalk-root: bulbous=b,club=c,cup=u,equal=e,rhizomorphs=z,rooted=r,missing=?
ncode=True
alphabet=['b','c','u','e','z','r','?'] # set missing to zero position
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c11=i
ncode=False
if ncode:
raise Exception("Encoding failed in c11 missing data maybe?")
return(c11)
######################################################################
def c12(stringvalue):
#stalk-surface-above-ring: fibrous=f,scaly=y,silky=k,smooth=s
ncode=True
alphabet=['f','y','k','s','?'] # set missing to zero position
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c12=i
ncode=False
if ncode:
raise Exception("Encoding failed in c12 missing data maybe?")
return(c12)
######################################################################
def c13(stringvalue):
#stalk-surface-below-ring: fibrous=f,scaly=y,silky=k,smooth=s
ncode=True
alphabet=['f','y','k','s','?'] # set missing to zero position
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c13=i
ncode=False
if ncode:
raise Exception("Encoding failed in c13 missing data maybe?")
return(c13)
######################################################################
def c14(stringvalue):
#stalk-color-above-ring: brown=n,buff=b,cinnamon=c,gray=g,orange=o,pink=p,red=e,white=w,yellow=y
ncode=True
alphabet=['n','b','c','g','o','p','e','w','y'] # set missing to zero position
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c14=i
ncode=False
if ncode:
raise Exception("Encoding failed in c14 missing data maybe?")
return(c14)
######################################################################
def c15(stringvalue):
#stalk-color-below-ring: brown=n,buff=b,cinnamon=c,gray=g,orange=o,pink=p,red=e,white=w,yellow=y
ncode=True
alphabet=['n','b','c','g','o','p','e','w','y'] # set missing to zero position
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c15=i
ncode=False
if ncode:
raise Exception("Encoding failed in c15 missing data maybe?")
return(c15)
######################################################################
def c16(stringvalue):
#veil-type: partial=p,universal=u
ncode=True
alphabet=['p','u'] #
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c16=i
ncode=False
if ncode:
raise Exception("Encoding failed in c16 missing data maybe?")
return(c16)
######################################################################
def c17(stringvalue):
#veil-color: brown=n,orange=o,white=w,yellow=y
ncode=True
alphabet=['n','o','w','y'] #
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c17=i
ncode=False
if ncode:
raise Exception("Encoding failed in c17 missing data maybe?")
return(c17)
######################################################################
def c18(stringvalue):
#ring-number: none=n,one=o,two=t
ncode=True
alphabet=['n','o','t'] #
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c18=i
ncode=False
if ncode:
raise Exception("Encoding failed in c18 missing data maybe?")
return(c18)
######################################################################
def c19(stringvalue):
#ring-type: cobwebby=c,evanescent=e,flaring=f,large=l,none=n,pendant=p,sheathing=s,zone=z
ncode=True
alphabet=['c','e','f','l','n','p','s','z'] #
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c19=i
ncode=False
if ncode:
raise Exception("Encoding failed in c19 missing data maybe?")
return(c19)
######################################################################
def c20(stringvalue):
#spore-print-color: black=k,brown=n,buff=b,chocolate=h,green=r,orange=o,purple=u,white=w,yellow=y
ncode=True
alphabet=['k','n','b','h','r','o','u','w','y','?'] #
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c20=i
ncode=False
if ncode:
raise Exception("Encoding failed in c20 missing data maybe?")
return(c20)
######################################################################
def c21(stringvalue):
#population:abundant=a,clustered=c,numerous=n,scattered=s,several=v,solitary=y
ncode=True
alphabet=['a','c','n','s','v','y','?'] #
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c21=i
ncode=False
if ncode:
raise Exception("Encoding failed in c21 missing data maybe?")
return(c21)
######################################################################
def c22(stringvalue):
# habitat: grasses=g,leaves=l,meadows=m,paths=p,urban=u,waste=w,woods=d
ncode=True
alphabet=['g','l','m','p','u','w','d']
for i in range(len(alphabet)):
if stringvalue == alphabet[i]:
c22=i
ncode=False
if ncode:
raise Exception("Encoding failed in c22 missing data maybe?")
return(c22)
interim['poison-class'] = mymushroom['poison-class'].apply(p0)
interim['cap-shape'] = mymushroom['cap-shape'].apply(c1)
interim['cap-surface'] = mymushroom['cap-surface'].apply(c2)
interim['cap-color'] = mymushroom['cap-color'].apply(c3)
interim['bruises'] = mymushroom['bruises'].apply(c4)
interim['odor'] = mymushroom['odor'].apply(c5)
interim['gill-attachment'] = mymushroom['gill-attachment'].apply(c6)
interim['gill-spacing'] = mymushroom['gill-spacing'].apply(c7)
interim['gill-size'] = mymushroom['gill-size'].apply(c8)
interim['gill-color'] = mymushroom['gill-color'].apply(c9)
interim['stalk-shape'] = mymushroom['stalk-shape'].apply(c10)
interim['stalk-root'] = mymushroom['stalk-root'].apply(c11)
interim['stalk-surface-above-ring']= mymushroom['stalk-surface-above-ring'].apply(c12)
interim['stalk-surface-below-ring']= mymushroom['stalk-surface-below-ring'].apply(c13)
interim['stalk-color-above-ring'] = mymushroom['stalk-color-above-ring'].apply(c14)
interim['stalk-color-below-ring'] = mymushroom['stalk-color-below-ring'].apply(c15)
interim['veil-type'] = mymushroom['veil-type'].apply(c16)
interim['veil-color'] = mymushroom['veil-color'].apply(c17)
interim['ring-number'] = mymushroom['ring-number'].apply(c18)
interim['ring-type'] = mymushroom['ring-type'].apply(c19)
interim['spore-print-color'] = mymushroom['spore-print-color'].apply(c20)
interim['population'] = mymushroom['population'].apply(c21)
interim['habitat'] = mymushroom['habitat'].apply(c22)
interim.head()
| poison-class | cap-shape | cap-surface | cap-color | bruises | odor | gill-attachment | gill-spacing | gill-size | gill-color | ... | stalk-surface-below-ring | stalk-color-above-ring | stalk-color-below-ring | veil-type | veil-color | ring-number | ring-type | spore-print-color | population | habitat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 0 | 1 | 7 | 2 | 0 | 1 | 0 | ... | 3 | 7 | 7 | 0 | 2 | 1 | 5 | 0 | 3 | 4 |
| 1 | 0 | 2 | 3 | 9 | 1 | 0 | 2 | 0 | 0 | 0 | ... | 3 | 7 | 7 | 0 | 2 | 1 | 5 | 1 | 2 | 0 |
| 2 | 0 | 0 | 3 | 8 | 1 | 1 | 2 | 0 | 0 | 1 | ... | 3 | 7 | 7 | 0 | 2 | 1 | 5 | 1 | 2 | 2 |
| 3 | 1 | 2 | 2 | 8 | 1 | 7 | 2 | 0 | 1 | 1 | ... | 3 | 7 | 7 | 0 | 2 | 1 | 5 | 0 | 3 | 4 |
| 4 | 0 | 2 | 3 | 3 | 0 | 6 | 2 | 1 | 0 | 0 | ... | 3 | 7 | 7 | 0 | 2 | 1 | 1 | 1 | 0 | 0 |
5 rows × 23 columns
#split dataset in features and target variable
#feature_cols = ['cap-shape', 'cap-surface','cap-color','bruises','odor','gill-attachment','gill-size','gill-color','habitat']
feature_cols = attributes[1:]
#feature_cols = attributes[1:16]+attributes[17:]
X = interim[feature_cols] # Features
y = interim['poison-class'] # Target variable
# split X and y into training and testing sets
from sklearn.model_selection import train_test_split
X_train,X_test,y_train,y_test=train_test_split(X,y,test_size=0.25,random_state=0)
feature_cols[16]
'veil-color'
# import the class
from sklearn.linear_model import LogisticRegression
# instantiate the model (using the default parameters)
#logreg = LogisticRegression()
logreg = LogisticRegression(max_iter=1000)
# fit the model with data
logreg.fit(X_train,y_train)
#
y_pred=logreg.predict(X_test)
print(logreg.intercept_[0])
print(logreg.coef_)
#y.head()
7.985575946121393
[[-0.10794435 1.08926906 0.08797603 -3.47364929 0.14180387 0.1864682
-8.98042887 7.73751204 -0.11238355 -0.05928835 -2.02832314 -0.7033172
-0.05014807 -0.30924012 0.33874996 0. 0.31015366 -0.78437801
-0.84016796 0.46958621 -0.59083624 -0.196766 ]]
# import the metrics class
from sklearn import metrics
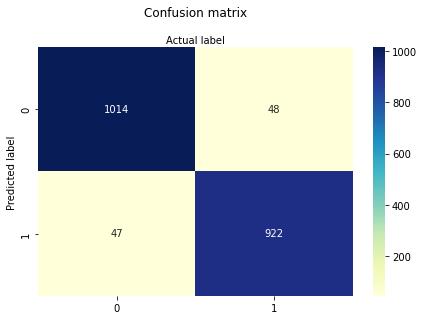
cnf_matrix = metrics.confusion_matrix(y_pred, y_test)
cnf_matrix
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
import pandas as pd
class_names=[0,1] # name of classes
fig, ax = plt.subplots()
tick_marks = np.arange(len(class_names))
plt.xticks(tick_marks, class_names)
plt.yticks(tick_marks, class_names)
# create heatmap
sns.heatmap(pd.DataFrame(cnf_matrix), annot=True, cmap="YlGnBu" ,fmt='g')
ax.xaxis.set_label_position("top")
plt.tight_layout()
plt.title('Confusion matrix', y=1.1)
plt.ylabel('Predicted label')
plt.xlabel('Actual label');
'''
/* Nonlinear Optimization using the algorithm of Hooke and Jeeves */
'''
'\n/* Nonlinear Optimization using the algorithm of Hooke and Jeeves */\n'
