Computational Thinking Concepts¶
The CT concepts expressed within Databases include:
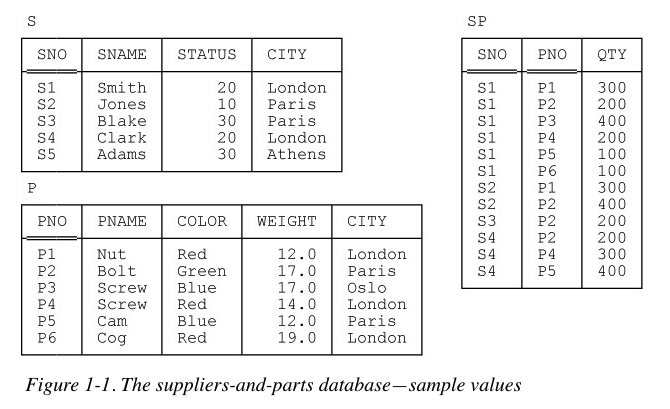
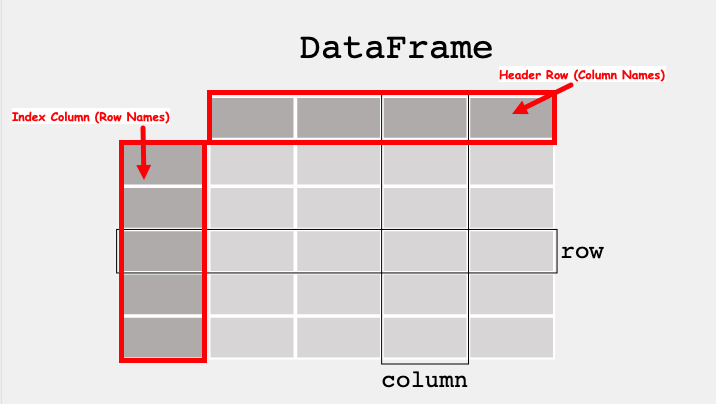
Decomposition: Break a problem down into smaller pieces; Collections of data are decomposed into their smallest usable partAbstraction: A database is an abstraction of a collection of data
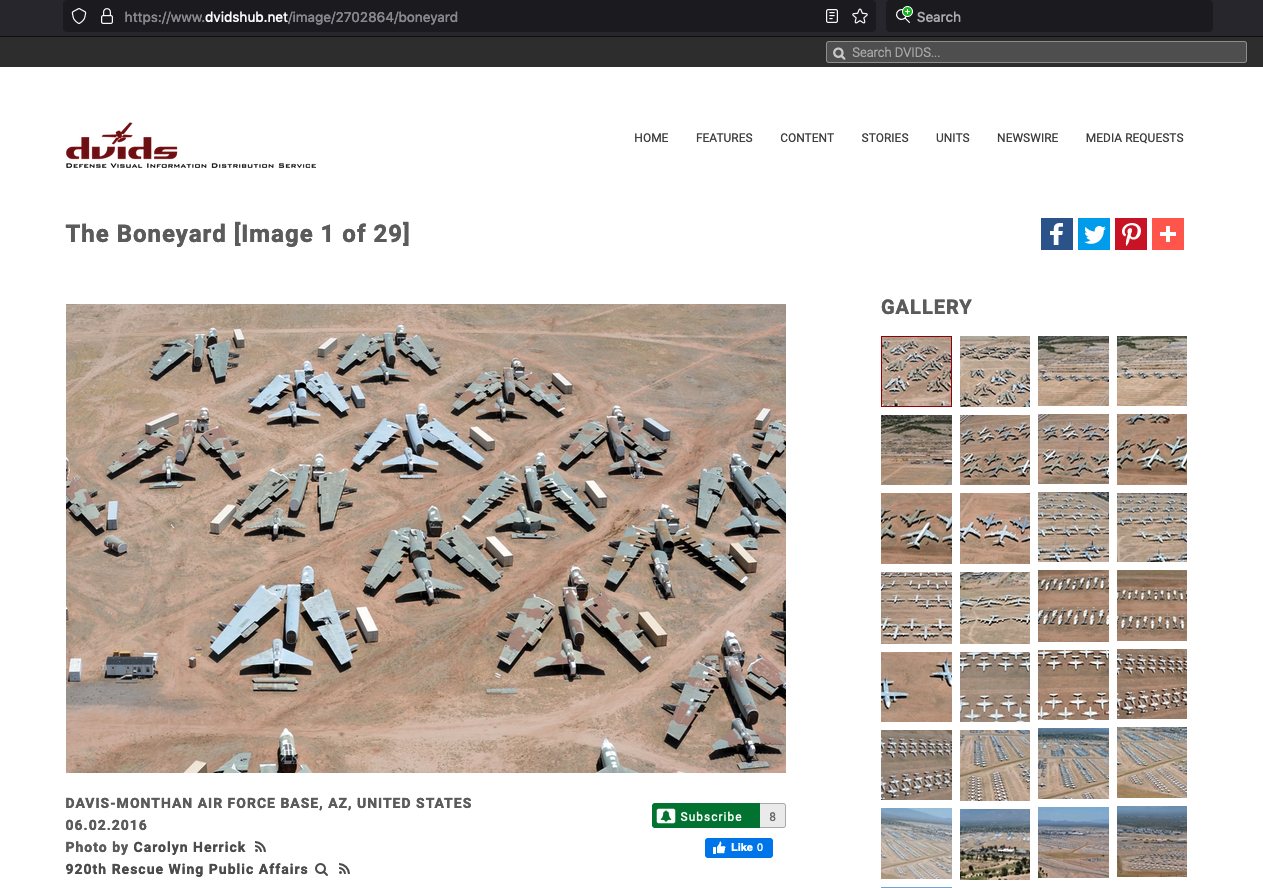
Suppose we need to store a car as a collection of its parts (implying dissassembly each time we park it), such decompotion would produce a situation like the image.

If the location of each part is recorded, then we can determine if something is missing as in

In the image, the two missing parts are pretty large, and would be evident on a fully assembled car (missing front corner panel, and right rear tire. Smaller parts would be harder to track on the fully assembled object. However if we had two fully assembled cars, and when we moved them heard the tink-tink-tink of a ball bearing bouncing on the floor, we would know something is missing - a query of the database to find where all the balls are supposed to be will help us figure out which car is incomplete.
In other contexts you wouldn’t want to have to take your car apart and store every piece separately whenever you park it in the garage. In that case, you would want to store your car as a single entry in the database (garage), and access its pieces through it (used car parts are usually sourced from fully assembled cars. The U.S. Airforce keeps a lot of otherwise broken aircraft for parts replacement. As a part is removed it is entered into a database "a transaction" so they know that part is no longer in the broken aircraft lot but in service somewhere. So the database may locate a part in a bin in a hangar or a part that is residing in an assembled aircraft. In either case, the hangar (and parts bin) as well as the broken aircarft are both within the database schema - an abstraction.